Benchmarks — Detailed Results¶
In one of the above sections,
we have summarised the AR indices based on the datasets from
the Benchmark Suite for Clustering Algorithms (Version 1.0)
[12].
In this section, we present more detailed results for
some other partition similarity measures implemented in the genieclust
package: Fowlkes–Mallows [8], adjusted Rand [20],
adjusted and normalised mutual information [34],
normalised pivoted accuracy (which is based on set-matching classification rate),
normalised clustering accuracy [13],
and pair sets index [32]; for more details,
see the API documentation of genieclust.compare_partitions.
In each case, a score of 1.0 denotes perfect agreement between the clustering
results and the reference partitions.
At the preprocessing stage, features with variance of 0 were removed. Every dataset has been centred at 0 and scaled so that is has total variance of 1. Then, a tiny bit of Gaussian noise has been added to each item. Clustering is performed with respect to the Euclidean distance (wherever applicable).
All raw results can be found here.
Small Datasets¶
nca¶
Summary statistics for nca (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.6 |
0.34 |
0 |
0.34 |
0.64 |
0.96 |
1 |
Birch_0.01 |
73 |
0.67 |
0.3 |
0.01 |
0.44 |
0.76 |
0.96 |
1 |
Complete linkage |
73 |
0.6 |
0.3 |
0.02 |
0.41 |
0.57 |
0.83 |
1 |
Gaussian mixtures |
73 |
0.72 |
0.32 |
0.01 |
0.53 |
0.85 |
0.99 |
1 |
Genie_0.1 |
73 |
0.81 |
0.24 |
0 |
0.7 |
0.93 |
1 |
1 |
Genie_0.3 |
73 |
0.82 |
0.24 |
0 |
0.64 |
0.95 |
1 |
1 |
Genie_0.5 |
73 |
0.79 |
0.26 |
0.15 |
0.63 |
0.94 |
1 |
1 |
ITM |
73 |
0.77 |
0.22 |
0.09 |
0.67 |
0.8 |
0.99 |
1 |
K-means |
73 |
0.67 |
0.3 |
0.01 |
0.46 |
0.7 |
0.97 |
1 |
Single linkage |
73 |
0.44 |
0.43 |
0 |
0.01 |
0.3 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.69 |
0.34 |
0 |
0.44 |
0.84 |
1 |
1 |
Ward linkage |
73 |
0.67 |
0.29 |
0.05 |
0.44 |
0.78 |
0.95 |
1 |
Ranks for nca (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.7 |
3.7 |
1 |
4 |
8 |
10 |
12 |
Birch_0.01 |
72 |
5.5 |
3.2 |
1 |
2 |
6 |
8 |
12 |
Complete linkage |
72 |
7.8 |
2.8 |
1 |
7 |
8 |
10 |
12 |
Gaussian mixtures |
72 |
4.6 |
3.9 |
1 |
1 |
3.5 |
7.2 |
12 |
Genie_0.1 |
72 |
3.5 |
3.3 |
1 |
1 |
2 |
4.2 |
11 |
Genie_0.3 |
72 |
3.8 |
3.5 |
1 |
1 |
2 |
7 |
12 |
Genie_0.5 |
72 |
4.4 |
4 |
1 |
1 |
2 |
9 |
11 |
ITM |
72 |
5.1 |
3.7 |
1 |
1 |
5 |
8.2 |
12 |
K-means |
72 |
5.2 |
3.6 |
1 |
1 |
5.5 |
8 |
12 |
Single linkage |
72 |
8.1 |
5 |
1 |
1 |
12 |
12 |
12 |
Spectral_RBF_5 |
72 |
5.4 |
3.8 |
1 |
1 |
6 |
9 |
12 |
Ward linkage |
72 |
5.8 |
3.4 |
1 |
2.8 |
6 |
8 |
12 |
Raw results for nca (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.31 |
0.31 |
0.29 |
0.17 |
1 |
1 |
1 |
1 |
0.43 |
1 |
1 |
0.31 |
fcps/chainlink |
0.52 |
0.53 |
0.56 |
0.95 |
1 |
1 |
1 |
1 |
0.31 |
1 |
1 |
0.53 |
fcps/engytime |
0.23 |
0.88 |
0.21 |
0.99 |
0.92 |
0.92 |
0.92 |
0.91 |
0.92 |
0 |
0.96 |
0.86 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.94 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.61 |
0.66 |
0.66 |
1 |
1 |
1 |
1 |
1 |
0.7 |
1 |
0.86 |
0.64 |
fcps/target |
0.86 |
0.84 |
0.88 |
0.84 |
1 |
1 |
1 |
1 |
0.84 |
1 |
1 |
0.83 |
fcps/tetra |
1 |
0.98 |
0.99 |
1 |
1 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.98 |
fcps/twodiamonds |
1 |
1 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.93 |
1 |
1 |
1 |
1 |
0.93 |
1 |
0.88 |
0.78 |
graves/dense |
0.96 |
0.96 |
0.51 |
1 |
0.98 |
0.98 |
0.98 |
1 |
0.9 |
0.03 |
0.96 |
0.96 |
graves/fuzzyx |
0.86 |
0.95 |
0.97 |
1 |
0.84 |
0.72 |
0.63 |
0.97 |
1 |
0 |
0.78 |
0.88 |
graves/line |
0.37 |
0.37 |
0.36 |
1 |
0.5 |
1 |
1 |
0.6 |
0.46 |
1 |
1 |
0.25 |
graves/parabolic |
0.77 |
0.76 |
0.77 |
0.73 |
0.9 |
0.9 |
0.9 |
0.8 |
0.77 |
0 |
0.81 |
0.79 |
graves/ring |
0.34 |
0.34 |
0.46 |
0.17 |
1 |
1 |
1 |
1 |
0.01 |
1 |
1 |
0.34 |
graves/ring_noisy |
0 |
0.34 |
0.52 |
0.16 |
1 |
1 |
1 |
1 |
0.41 |
0 |
1 |
0.39 |
graves/ring_outliers |
0.58 |
0.33 |
0.56 |
0.31 |
1 |
1 |
1 |
1 |
0.57 |
1 |
1 |
0.34 |
graves/zigzag |
0.69 |
0.74 |
0.57 |
0.98 |
1 |
1 |
1 |
0.87 |
0.7 |
1 |
0.8 |
0.64 |
graves/zigzag_noisy |
0.65 |
0.77 |
0.58 |
0.95 |
0.88 |
1 |
1 |
0.75 |
0.64 |
0.5 |
0.78 |
0.79 |
graves/zigzag_outliers |
0.5 |
0.68 |
0.57 |
0.99 |
1 |
1 |
1 |
0.91 |
0.62 |
0.5 |
0.4 |
0.62 |
other/chameleon_t4_8k |
0.64 |
0.62 |
0.53 |
0.59 |
0.72 |
1 |
0.8 |
0.78 |
0.61 |
0 |
0.61 |
0.58 |
other/chameleon_t5_8k |
1 |
1 |
0.8 |
1 |
1 |
1 |
0.8 |
0.69 |
1 |
0 |
1 |
1 |
other/chameleon_t7_10k |
0.59 |
0.62 |
0.56 |
0.59 |
0.73 |
0.87 |
1 |
0.75 |
0.54 |
0 |
0.55 |
0.64 |
other/chameleon_t8_8k |
0.45 |
0.49 |
0.48 |
0.59 |
0.67 |
0.48 |
0.73 |
0.62 |
0.52 |
0 |
0.5 |
0.48 |
other/hdbscan |
0.38 |
0.69 |
0.52 |
0.83 |
0.98 |
0.78 |
0.79 |
0.89 |
0.77 |
0 |
0.45 |
0.9 |
other/iris |
0.86 |
0.84 |
0.76 |
0.95 |
0.94 |
0.94 |
0.55 |
0.94 |
0.84 |
0.52 |
0.85 |
0.84 |
other/iris5 |
0.86 |
0.84 |
0.76 |
0.95 |
0.41 |
0.41 |
0.94 |
0.28 |
0.84 |
0.52 |
0.85 |
0.84 |
other/square |
0.39 |
0.01 |
0.41 |
0.19 |
1 |
1 |
1 |
1 |
0.17 |
1 |
0.41 |
0.5 |
sipu/a1 |
0.96 |
0.96 |
0.96 |
0.98 |
0.97 |
0.91 |
0.84 |
0.82 |
0.98 |
0.37 |
0.97 |
0.95 |
sipu/a2 |
0.97 |
0.97 |
0.95 |
0.98 |
0.97 |
0.94 |
0.83 |
0.83 |
0.98 |
0.3 |
0.93 |
0.96 |
sipu/a3 |
0.97 |
0.97 |
0.96 |
0.96 |
0.98 |
0.95 |
0.84 |
0.83 |
0.96 |
0.25 |
0.95 |
0.97 |
sipu/aggregation |
1 |
0.78 |
0.75 |
1 |
0.58 |
0.58 |
0.88 |
0.66 |
0.75 |
0.66 |
1 |
0.78 |
sipu/compound |
0.67 |
0.87 |
0.67 |
0.84 |
0.84 |
0.87 |
0.76 |
0.79 |
0.81 |
0.67 |
0.77 |
0.87 |
sipu/d31 |
0.94 |
0.96 |
0.96 |
0.97 |
0.97 |
0.93 |
0.76 |
0.85 |
0.98 |
0.24 |
0.97 |
0.96 |
sipu/flame |
0.74 |
0.59 |
0.21 |
0.68 |
1 |
1 |
1 |
0.68 |
0.74 |
0.02 |
0.91 |
0.59 |
sipu/jain |
0.79 |
0.81 |
0.79 |
0.43 |
0.49 |
1 |
1 |
0.71 |
0.7 |
0.27 |
0.81 |
0.81 |
sipu/pathbased |
0.63 |
0.67 |
0.56 |
0.61 |
0.98 |
0.98 |
0.76 |
0.68 |
0.65 |
0.01 |
0.65 |
0.67 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.99 |
0.99 |
0.98 |
0.99 |
0.99 |
0.99 |
0.99 |
0.8 |
0.99 |
0.43 |
0.99 |
0.99 |
sipu/s2 |
0.95 |
0.95 |
0.83 |
0.97 |
0.95 |
0.95 |
0.83 |
0.86 |
0.97 |
0 |
0.97 |
0.95 |
sipu/s3 |
0.66 |
0.8 |
0.57 |
0.85 |
0.82 |
0.76 |
0.63 |
0.76 |
0.84 |
0 |
0.84 |
0.81 |
sipu/s4 |
0.59 |
0.68 |
0.52 |
0.79 |
0.77 |
0.73 |
0.54 |
0.67 |
0.78 |
0 |
0.69 |
0.68 |
sipu/spiral |
0.04 |
0.07 |
0.06 |
0.03 |
1 |
1 |
1 |
0.83 |
0.01 |
1 |
0.02 |
0.07 |
sipu/unbalance |
1 |
1 |
0.8 |
1 |
0.24 |
0.29 |
0.35 |
0.22 |
1 |
0.86 |
1 |
1 |
uci/ecoli |
0.5 |
0.52 |
0.47 |
0.51 |
0.42 |
0.44 |
0.4 |
0.46 |
0.57 |
0.2 |
0.48 |
0.52 |
uci/glass |
0.08 |
0.31 |
0.28 |
0.3 |
0.39 |
0.42 |
0.32 |
0.43 |
0.37 |
0.06 |
0.23 |
0.34 |
uci/ionosphere |
0.01 |
0.44 |
0.02 |
0.65 |
0.41 |
0.41 |
0.15 |
0.36 |
0.44 |
0.01 |
0.01 |
0.44 |
uci/sonar |
0.05 |
0.05 |
0.04 |
0.09 |
0 |
0 |
0.16 |
0.09 |
0.11 |
0.01 |
0.01 |
0.05 |
uci/statlog |
0 |
0.45 |
0.17 |
0.53 |
0.74 |
0.59 |
0.51 |
0.6 |
0.43 |
0 |
nan |
0.41 |
uci/wdbc |
0.09 |
0.41 |
0.09 |
0.8 |
0.45 |
0.4 |
0.4 |
0.8 |
0.61 |
0 |
0 |
0.41 |
uci/wine |
0.34 |
0.53 |
0.48 |
0.92 |
0.58 |
0.58 |
0.22 |
0.57 |
0.54 |
0.04 |
0.36 |
0.53 |
uci/yeast |
0.19 |
0.37 |
0.3 |
0.32 |
0.34 |
0.33 |
0.28 |
0.33 |
0.38 |
0.14 |
0.16 |
0.35 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.03 |
0.17 |
0.32 |
0.48 |
0.87 |
0.68 |
0.38 |
0.44 |
0.39 |
0 |
0.01 |
0.31 |
wut/graph |
0.56 |
0.58 |
0.56 |
0.93 |
0.63 |
0.59 |
0.45 |
0.63 |
0.59 |
0.07 |
0.58 |
0.62 |
wut/isolation |
0.03 |
0.04 |
0.02 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.05 |
wut/labirynth |
0.59 |
0.8 |
0.7 |
0.66 |
0.74 |
0.62 |
0.71 |
0.7 |
0.56 |
0.6 |
0.56 |
0.55 |
wut/mk1 |
1 |
1 |
0.98 |
1 |
1 |
1 |
1 |
0.68 |
1 |
0.5 |
1 |
1 |
wut/mk2 |
0.07 |
0.11 |
0.09 |
0.09 |
1 |
1 |
1 |
1 |
0.09 |
1 |
0.08 |
0.07 |
wut/mk3 |
0.5 |
0.92 |
0.92 |
0.94 |
0.88 |
0.88 |
0.58 |
0.68 |
0.94 |
0 |
0.94 |
0.93 |
wut/mk4 |
0.18 |
0.31 |
0.42 |
0.55 |
1 |
1 |
1 |
0.74 |
0.37 |
1 |
0.39 |
0.36 |
wut/olympic |
0.27 |
0.26 |
0.21 |
0.2 |
0.32 |
0.29 |
0.25 |
0.36 |
0.22 |
0 |
0.26 |
0.21 |
wut/smile |
0.99 |
0.86 |
0.63 |
0.5 |
0.55 |
1 |
1 |
0.7 |
0.86 |
1 |
1 |
0.88 |
wut/stripes |
0.05 |
0.04 |
0.11 |
0.11 |
1 |
1 |
1 |
1 |
0.11 |
1 |
0.11 |
0.12 |
wut/trajectories |
1 |
1 |
0.74 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.58 |
0.61 |
0.72 |
0.61 |
0.76 |
1 |
1 |
0.76 |
0.64 |
1 |
0.93 |
0.65 |
wut/twosplashes |
0.05 |
0.53 |
0.47 |
0.82 |
0.71 |
0.71 |
0.71 |
0.86 |
0.53 |
0.01 |
0.53 |
0.44 |
wut/windows |
0.8 |
0.8 |
0.32 |
0.82 |
0.57 |
0.57 |
1 |
0.36 |
0.69 |
1 |
0.3 |
0.79 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.65 |
0.98 |
1 |
0.87 |
0.87 |
0.87 |
0.59 |
0.87 |
0.98 |
0.01 |
0.34 |
0.98 |
wut/x3 |
0.94 |
0.99 |
0.74 |
0.96 |
0.93 |
0.94 |
0.94 |
0.6 |
1 |
0.03 |
0.54 |
0.99 |
wut/z1 |
0.32 |
0.27 |
0.36 |
0.11 |
0.5 |
0.5 |
0.3 |
0.5 |
0.31 |
0.06 |
0.33 |
0.27 |
wut/z2 |
0.75 |
0.88 |
0.73 |
1 |
0.88 |
0.64 |
0.64 |
0.88 |
0.86 |
0.5 |
0.99 |
0.84 |
wut/z3 |
1 |
0.99 |
0.95 |
0.99 |
0.7 |
0.49 |
0.94 |
0.83 |
1 |
0.66 |
0.98 |
1 |
ar¶
Summary statistics for ar (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.51 |
0.38 |
0 |
0.11 |
0.51 |
0.93 |
1 |
Birch_0.01 |
73 |
0.55 |
0.36 |
0 |
0.22 |
0.56 |
0.93 |
1 |
Complete linkage |
73 |
0.48 |
0.34 |
0 |
0.21 |
0.4 |
0.79 |
1 |
Gaussian mixtures |
73 |
0.65 |
0.37 |
0 |
0.4 |
0.82 |
0.98 |
1 |
Genie_0.1 |
73 |
0.74 |
0.31 |
0 |
0.53 |
0.88 |
1 |
1 |
Genie_0.3 |
73 |
0.78 |
0.27 |
0 |
0.59 |
0.94 |
1 |
1 |
Genie_0.5 |
73 |
0.77 |
0.3 |
0 |
0.66 |
0.92 |
1 |
1 |
ITM |
73 |
0.68 |
0.27 |
0 |
0.53 |
0.69 |
0.99 |
1 |
K-means |
73 |
0.55 |
0.35 |
0 |
0.2 |
0.51 |
0.95 |
1 |
Single linkage |
73 |
0.44 |
0.45 |
0 |
0 |
0.32 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.63 |
0.37 |
0 |
0.33 |
0.73 |
0.99 |
1 |
Ward linkage |
73 |
0.54 |
0.35 |
0 |
0.22 |
0.54 |
0.91 |
1 |
Ranks for ar (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.6 |
3.5 |
1 |
4.8 |
7 |
9.2 |
12 |
Birch_0.01 |
72 |
5.8 |
2.9 |
1 |
4 |
6 |
8 |
12 |
Complete linkage |
72 |
7.7 |
3.2 |
1 |
6 |
8 |
11 |
12 |
Gaussian mixtures |
72 |
4.2 |
3.6 |
1 |
1 |
3 |
7 |
12 |
Genie_0.1 |
72 |
3.8 |
3.3 |
1 |
1 |
3 |
6 |
12 |
Genie_0.3 |
72 |
3.3 |
3 |
1 |
1 |
2 |
5 |
11 |
Genie_0.5 |
72 |
4.2 |
3.9 |
1 |
1 |
2 |
8 |
11 |
ITM |
72 |
5.4 |
3.9 |
1 |
1 |
5 |
9 |
12 |
K-means |
72 |
5.6 |
3.8 |
1 |
1 |
6 |
9 |
12 |
Single linkage |
72 |
7.4 |
5.1 |
1 |
1 |
11 |
12 |
12 |
Spectral_RBF_5 |
72 |
5.2 |
3.5 |
1 |
1 |
6 |
8 |
11 |
Ward linkage |
72 |
6 |
3 |
1 |
4 |
6 |
8 |
12 |
Raw results for ar (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.1 |
0.1 |
0.08 |
0.03 |
1 |
1 |
1 |
1 |
0.18 |
1 |
1 |
0.1 |
fcps/chainlink |
0.27 |
0.28 |
0.31 |
0.91 |
1 |
1 |
1 |
1 |
0.09 |
1 |
1 |
0.28 |
fcps/engytime |
0.05 |
0.78 |
0.04 |
0.98 |
0.84 |
0.84 |
0.84 |
0.83 |
0.85 |
0 |
0.92 |
0.75 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.9 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.36 |
0.4 |
0.4 |
1 |
1 |
1 |
1 |
1 |
0.44 |
1 |
0.8 |
0.37 |
fcps/target |
0.15 |
0.64 |
0.21 |
0.65 |
1 |
1 |
1 |
1 |
0.63 |
1 |
1 |
0.64 |
fcps/tetra |
0.99 |
0.97 |
0.99 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0.97 |
fcps/twodiamonds |
0.99 |
0.99 |
0.99 |
1 |
0.98 |
0.98 |
0.98 |
0.99 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.86 |
1 |
1 |
1 |
1 |
0.86 |
1 |
0.78 |
0.6 |
graves/dense |
0.92 |
0.92 |
0.26 |
1 |
0.96 |
0.96 |
0.96 |
1 |
0.81 |
0 |
0.92 |
0.92 |
graves/fuzzyx |
0.74 |
0.9 |
0.94 |
1 |
0.73 |
0.5 |
0.4 |
0.95 |
1 |
0 |
0.59 |
0.78 |
graves/line |
0 |
0 |
0 |
1 |
0.02 |
1 |
1 |
0.12 |
0 |
1 |
1 |
0 |
graves/parabolic |
0.6 |
0.57 |
0.59 |
0.54 |
0.81 |
0.81 |
0.81 |
0.64 |
0.59 |
0 |
0.66 |
0.62 |
graves/ring |
0.11 |
0.12 |
0.21 |
0.03 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
0.12 |
graves/ring_noisy |
0 |
0.11 |
0.27 |
0.02 |
1 |
1 |
1 |
1 |
0.16 |
0 |
1 |
0.15 |
graves/ring_outliers |
0.63 |
0.63 |
0.34 |
0.62 |
1 |
1 |
1 |
1 |
0.62 |
1 |
1 |
0.63 |
graves/zigzag |
0.53 |
0.62 |
0.36 |
0.96 |
1 |
1 |
1 |
0.78 |
0.53 |
1 |
0.68 |
0.54 |
graves/zigzag_noisy |
0.52 |
0.64 |
0.52 |
0.9 |
0.77 |
1 |
1 |
0.54 |
0.51 |
0.47 |
0.63 |
0.66 |
graves/zigzag_outliers |
0.31 |
0.51 |
0.33 |
0.98 |
1 |
1 |
1 |
0.83 |
0.44 |
0.48 |
0.34 |
0.39 |
other/chameleon_t4_8k |
0.64 |
0.62 |
0.55 |
0.56 |
0.83 |
1 |
0.93 |
0.84 |
0.6 |
0 |
0.63 |
0.61 |
other/chameleon_t5_8k |
1 |
1 |
0.73 |
1 |
1 |
1 |
0.83 |
0.59 |
1 |
0 |
1 |
1 |
other/chameleon_t7_10k |
0.45 |
0.44 |
0.37 |
0.4 |
0.53 |
0.7 |
1 |
0.53 |
0.42 |
0 |
0.38 |
0.43 |
other/chameleon_t8_8k |
0.37 |
0.39 |
0.33 |
0.44 |
0.61 |
0.64 |
0.71 |
0.57 |
0.37 |
0 |
0.36 |
0.37 |
other/hdbscan |
0.43 |
0.63 |
0.46 |
0.82 |
0.97 |
0.71 |
0.71 |
0.75 |
0.64 |
0 |
0.33 |
0.84 |
other/iris |
0.76 |
0.73 |
0.64 |
0.9 |
0.89 |
0.89 |
0.56 |
0.89 |
0.73 |
0.56 |
0.75 |
0.73 |
other/iris5 |
0.56 |
0.51 |
0.34 |
0.82 |
0.59 |
0.59 |
0.79 |
0.52 |
0.51 |
0.15 |
0.53 |
0.51 |
other/square |
0.15 |
0 |
0.17 |
0.04 |
1 |
1 |
1 |
1 |
0.03 |
1 |
0.17 |
0.25 |
sipu/a1 |
0.93 |
0.93 |
0.92 |
0.96 |
0.94 |
0.9 |
0.83 |
0.77 |
0.97 |
0.44 |
0.94 |
0.91 |
sipu/a2 |
0.93 |
0.94 |
0.91 |
0.96 |
0.95 |
0.92 |
0.83 |
0.77 |
0.97 |
0.35 |
0.91 |
0.92 |
sipu/a3 |
0.94 |
0.94 |
0.92 |
0.95 |
0.96 |
0.94 |
0.82 |
0.77 |
0.95 |
0.32 |
0.93 |
0.94 |
sipu/aggregation |
1 |
0.82 |
0.78 |
1 |
0.48 |
0.57 |
0.88 |
0.61 |
0.76 |
0.8 |
0.99 |
0.81 |
sipu/compound |
0.91 |
0.88 |
0.91 |
0.91 |
0.78 |
0.78 |
0.88 |
0.62 |
0.76 |
0.93 |
0.87 |
0.88 |
sipu/d31 |
0.91 |
0.93 |
0.92 |
0.95 |
0.94 |
0.9 |
0.71 |
0.8 |
0.95 |
0.17 |
0.94 |
0.92 |
sipu/flame |
0.44 |
0.22 |
0 |
0.34 |
1 |
1 |
1 |
0.35 |
0.48 |
0.01 |
0.83 |
0.22 |
sipu/jain |
0.78 |
0.51 |
0.78 |
0 |
0.04 |
1 |
1 |
0.32 |
0.32 |
0.26 |
0.51 |
0.51 |
sipu/pathbased |
0.59 |
0.54 |
0.41 |
0.6 |
0.97 |
0.97 |
0.7 |
0.54 |
0.5 |
0 |
0.6 |
0.54 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.98 |
0.99 |
0.97 |
0.99 |
0.99 |
0.99 |
0.99 |
0.76 |
0.99 |
0.46 |
0.99 |
0.98 |
sipu/s2 |
0.91 |
0.9 |
0.79 |
0.94 |
0.92 |
0.92 |
0.78 |
0.77 |
0.94 |
0 |
0.94 |
0.91 |
sipu/s3 |
0.6 |
0.68 |
0.51 |
0.73 |
0.69 |
0.67 |
0.56 |
0.61 |
0.72 |
0 |
0.71 |
0.68 |
sipu/s4 |
0.49 |
0.56 |
0.44 |
0.64 |
0.62 |
0.59 |
0.47 |
0.55 |
0.63 |
0 |
0.57 |
0.55 |
sipu/spiral |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0.73 |
0 |
1 |
0 |
0 |
sipu/unbalance |
1 |
1 |
0.61 |
1 |
0.57 |
0.62 |
0.78 |
0.53 |
1 |
1 |
1 |
1 |
uci/ecoli |
0.74 |
0.49 |
0.62 |
0.61 |
0.36 |
0.46 |
0.66 |
0.33 |
0.46 |
0.04 |
0.35 |
0.49 |
uci/glass |
0.02 |
0.25 |
0.23 |
0.24 |
0.12 |
0.25 |
0.22 |
0.23 |
0.27 |
0.01 |
0.22 |
0.26 |
uci/ionosphere |
0 |
0.19 |
0.01 |
0.4 |
0.21 |
0.21 |
0 |
0.09 |
0.18 |
0 |
0 |
0.19 |
uci/sonar |
0.01 |
0 |
0 |
0 |
0 |
0 |
0.01 |
0 |
0.01 |
0 |
0 |
0 |
uci/statlog |
0 |
0.33 |
0.1 |
0.47 |
0.62 |
0.52 |
0.47 |
0.53 |
0.36 |
0 |
nan |
0.31 |
uci/wdbc |
0.05 |
0.29 |
0.05 |
0.71 |
0.09 |
0.28 |
0.28 |
0.63 |
0.49 |
0 |
0 |
0.29 |
uci/wine |
0.29 |
0.37 |
0.37 |
0.82 |
0.36 |
0.36 |
0.25 |
0.39 |
0.37 |
0.01 |
0.32 |
0.37 |
uci/yeast |
0.01 |
0.12 |
0.09 |
0.05 |
0.11 |
0.18 |
0.08 |
0.08 |
0.14 |
0.01 |
0.01 |
0.13 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0 |
0.02 |
0.36 |
0.47 |
0.76 |
0.46 |
0.1 |
0.53 |
0.11 |
0 |
0 |
0.07 |
wut/graph |
0.41 |
0.44 |
0.4 |
0.86 |
0.48 |
0.49 |
0.39 |
0.52 |
0.44 |
0.03 |
0.41 |
0.46 |
wut/isolation |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
wut/labirynth |
0.36 |
0.49 |
0.3 |
0.62 |
0.5 |
0.59 |
0.72 |
0.72 |
0.29 |
0.76 |
0.39 |
0.34 |
wut/mk1 |
0.99 |
0.99 |
0.97 |
0.99 |
0.99 |
0.99 |
0.99 |
0.53 |
0.99 |
0.56 |
0.99 |
0.99 |
wut/mk2 |
0 |
0.01 |
0.01 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0 |
0 |
wut/mk3 |
0.56 |
0.85 |
0.84 |
0.88 |
0.8 |
0.8 |
0.56 |
0.54 |
0.89 |
0 |
0.88 |
0.86 |
wut/mk4 |
0.04 |
0.14 |
0.28 |
0.5 |
1 |
1 |
1 |
0.59 |
0.2 |
1 |
0.2 |
0.19 |
wut/olympic |
0.14 |
0.15 |
0.12 |
0.14 |
0.17 |
0.15 |
0.09 |
0.21 |
0.11 |
0 |
0.13 |
0.13 |
wut/smile |
0.99 |
0.61 |
0.77 |
0.61 |
0.64 |
1 |
1 |
0.62 |
0.61 |
1 |
1 |
0.65 |
wut/stripes |
0 |
0 |
0.01 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.01 |
wut/trajectories |
1 |
1 |
0.71 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.13 |
0.14 |
0.25 |
0.14 |
0.39 |
1 |
1 |
0.39 |
0.15 |
1 |
0.75 |
0.16 |
wut/twosplashes |
0 |
0.28 |
0.22 |
0.67 |
0.5 |
0.5 |
0.5 |
0.73 |
0.28 |
0 |
0.28 |
0.19 |
wut/windows |
0.1 |
0.1 |
0.09 |
0.14 |
0.14 |
0.2 |
1 |
0.35 |
0.09 |
1 |
0.15 |
0.1 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.51 |
0.97 |
1 |
0.69 |
0.81 |
0.81 |
0.5 |
0.69 |
0.97 |
0 |
0.27 |
0.97 |
wut/x3 |
0.96 |
0.98 |
0.47 |
0.93 |
0.88 |
0.96 |
0.96 |
0.65 |
1 |
0.02 |
0.51 |
0.98 |
wut/z1 |
0.21 |
0.1 |
0.19 |
0.01 |
0.39 |
0.39 |
0.14 |
0.39 |
0.2 |
0 |
0.15 |
0.1 |
wut/z2 |
0.51 |
0.5 |
0.44 |
1 |
0.5 |
0.63 |
0.82 |
0.49 |
0.47 |
0.73 |
0.99 |
0.43 |
wut/z3 |
1 |
1 |
0.93 |
1 |
0.63 |
0.66 |
0.92 |
0.65 |
1 |
0.74 |
0.98 |
1 |
fm¶
Summary statistics for fm (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.73 |
0.21 |
0.34 |
0.55 |
0.72 |
0.94 |
1 |
Birch_0.01 |
73 |
0.73 |
0.21 |
0.28 |
0.58 |
0.7 |
0.94 |
1 |
Complete linkage |
73 |
0.69 |
0.2 |
0.3 |
0.55 |
0.65 |
0.9 |
1 |
Gaussian mixtures |
73 |
0.79 |
0.22 |
0.26 |
0.66 |
0.87 |
0.99 |
1 |
Genie_0.1 |
73 |
0.82 |
0.2 |
0.25 |
0.66 |
0.92 |
1 |
1 |
Genie_0.3 |
73 |
0.85 |
0.18 |
0.37 |
0.73 |
0.94 |
1 |
1 |
Genie_0.5 |
73 |
0.86 |
0.18 |
0.36 |
0.75 |
0.94 |
1 |
1 |
ITM |
73 |
0.78 |
0.18 |
0.23 |
0.65 |
0.78 |
0.99 |
1 |
K-means |
73 |
0.72 |
0.22 |
0.29 |
0.51 |
0.7 |
0.95 |
1 |
Single linkage |
73 |
0.73 |
0.24 |
0.26 |
0.53 |
0.71 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.78 |
0.21 |
0.33 |
0.61 |
0.83 |
0.99 |
1 |
Ward linkage |
73 |
0.72 |
0.21 |
0.29 |
0.58 |
0.7 |
0.92 |
1 |
Ranks for fm (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
5.9 |
3.3 |
1 |
3 |
6 |
8.2 |
12 |
Birch_0.01 |
72 |
6 |
3 |
1 |
4.8 |
6.5 |
8 |
12 |
Complete linkage |
72 |
7.7 |
3.5 |
1 |
5.8 |
8 |
11 |
12 |
Gaussian mixtures |
72 |
4.5 |
3.6 |
1 |
1 |
3 |
8 |
12 |
Genie_0.1 |
72 |
4.1 |
3.6 |
1 |
1 |
3 |
7 |
12 |
Genie_0.3 |
72 |
3.5 |
3.1 |
1 |
1 |
2 |
5 |
11 |
Genie_0.5 |
72 |
3.9 |
3.6 |
1 |
1 |
2 |
6.2 |
12 |
ITM |
72 |
5.9 |
4.2 |
1 |
1 |
5 |
10 |
12 |
K-means |
72 |
6.3 |
4 |
1 |
1.8 |
8 |
9 |
12 |
Single linkage |
72 |
6.1 |
4.9 |
1 |
1 |
6 |
12 |
12 |
Spectral_RBF_5 |
72 |
4.8 |
3.4 |
1 |
1 |
5 |
8 |
11 |
Ward linkage |
72 |
6.3 |
3.1 |
1 |
5 |
7 |
9 |
12 |
Raw results for fm (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.65 |
0.65 |
0.65 |
0.66 |
1 |
1 |
1 |
1 |
0.65 |
1 |
1 |
0.65 |
fcps/chainlink |
0.68 |
0.68 |
0.69 |
0.95 |
1 |
1 |
1 |
1 |
0.55 |
1 |
1 |
0.68 |
fcps/engytime |
0.65 |
0.89 |
0.65 |
0.99 |
0.92 |
0.92 |
0.92 |
0.92 |
0.92 |
0.71 |
0.96 |
0.87 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.91 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.6 |
0.63 |
0.63 |
1 |
1 |
1 |
1 |
1 |
0.65 |
1 |
0.88 |
0.61 |
fcps/target |
0.71 |
0.8 |
0.71 |
0.8 |
1 |
1 |
1 |
1 |
0.79 |
1 |
1 |
0.79 |
fcps/tetra |
0.99 |
0.98 |
0.99 |
1 |
1 |
1 |
1 |
1 |
1 |
0.49 |
1 |
0.98 |
fcps/twodiamonds |
1 |
1 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0.71 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.93 |
1 |
1 |
1 |
1 |
0.93 |
1 |
0.89 |
0.8 |
graves/dense |
0.96 |
0.96 |
0.67 |
1 |
0.98 |
0.98 |
0.98 |
1 |
0.9 |
0.69 |
0.96 |
0.96 |
graves/fuzzyx |
0.8 |
0.95 |
0.97 |
1 |
0.8 |
0.66 |
0.66 |
0.96 |
1 |
0.71 |
0.7 |
0.89 |
graves/line |
0.6 |
0.6 |
0.61 |
1 |
0.6 |
1 |
1 |
0.63 |
0.6 |
1 |
1 |
0.64 |
graves/parabolic |
0.8 |
0.79 |
0.8 |
0.77 |
0.91 |
0.91 |
0.91 |
0.82 |
0.79 |
0.71 |
0.83 |
0.81 |
graves/ring |
0.65 |
0.65 |
0.66 |
0.66 |
1 |
1 |
1 |
1 |
0.5 |
1 |
1 |
0.65 |
graves/ring_noisy |
0.71 |
0.65 |
0.68 |
0.66 |
1 |
1 |
1 |
1 |
0.65 |
0.71 |
1 |
0.65 |
graves/ring_outliers |
0.78 |
0.78 |
0.65 |
0.78 |
1 |
1 |
1 |
1 |
0.78 |
1 |
1 |
0.78 |
graves/zigzag |
0.63 |
0.7 |
0.53 |
0.97 |
1 |
1 |
1 |
0.82 |
0.64 |
1 |
0.75 |
0.64 |
graves/zigzag_noisy |
0.62 |
0.71 |
0.63 |
0.92 |
0.82 |
1 |
1 |
0.63 |
0.62 |
0.67 |
0.72 |
0.73 |
graves/zigzag_outliers |
0.55 |
0.61 |
0.52 |
0.98 |
1 |
1 |
1 |
0.86 |
0.59 |
0.67 |
0.66 |
0.58 |
other/chameleon_t4_8k |
0.72 |
0.69 |
0.63 |
0.64 |
0.86 |
1 |
0.94 |
0.87 |
0.67 |
0.44 |
0.69 |
0.68 |
other/chameleon_t5_8k |
1 |
1 |
0.78 |
1 |
1 |
1 |
0.87 |
0.67 |
1 |
0.41 |
1 |
1 |
other/chameleon_t7_10k |
0.54 |
0.54 |
0.47 |
0.5 |
0.61 |
0.76 |
1 |
0.62 |
0.51 |
0.43 |
0.48 |
0.52 |
other/chameleon_t8_8k |
0.47 |
0.49 |
0.43 |
0.53 |
0.68 |
0.7 |
0.77 |
0.64 |
0.46 |
0.41 |
0.46 |
0.47 |
other/hdbscan |
0.6 |
0.7 |
0.58 |
0.85 |
0.97 |
0.79 |
0.79 |
0.8 |
0.7 |
0.42 |
0.53 |
0.87 |
other/iris |
0.84 |
0.82 |
0.77 |
0.94 |
0.92 |
0.92 |
0.75 |
0.92 |
0.82 |
0.76 |
0.83 |
0.82 |
other/iris5 |
0.77 |
0.74 |
0.67 |
0.9 |
0.76 |
0.76 |
0.89 |
0.72 |
0.73 |
0.69 |
0.75 |
0.74 |
other/square |
0.65 |
0.5 |
0.65 |
0.52 |
1 |
1 |
1 |
1 |
0.52 |
1 |
0.65 |
0.67 |
sipu/a1 |
0.93 |
0.94 |
0.92 |
0.96 |
0.94 |
0.9 |
0.85 |
0.78 |
0.97 |
0.56 |
0.94 |
0.92 |
sipu/a2 |
0.94 |
0.94 |
0.91 |
0.96 |
0.95 |
0.92 |
0.84 |
0.78 |
0.97 |
0.48 |
0.91 |
0.92 |
sipu/a3 |
0.94 |
0.94 |
0.92 |
0.95 |
0.96 |
0.94 |
0.84 |
0.77 |
0.95 |
0.45 |
0.93 |
0.94 |
sipu/aggregation |
1 |
0.86 |
0.83 |
1 |
0.58 |
0.66 |
0.91 |
0.69 |
0.82 |
0.86 |
0.99 |
0.86 |
sipu/compound |
0.94 |
0.92 |
0.94 |
0.94 |
0.85 |
0.85 |
0.92 |
0.74 |
0.83 |
0.95 |
0.91 |
0.92 |
sipu/d31 |
0.91 |
0.93 |
0.93 |
0.95 |
0.94 |
0.9 |
0.74 |
0.81 |
0.96 |
0.35 |
0.94 |
0.92 |
sipu/flame |
0.73 |
0.63 |
0.62 |
0.68 |
1 |
1 |
1 |
0.69 |
0.75 |
0.73 |
0.92 |
0.63 |
sipu/jain |
0.92 |
0.79 |
0.92 |
0.59 |
0.59 |
1 |
1 |
0.7 |
0.7 |
0.8 |
0.79 |
0.79 |
sipu/pathbased |
0.73 |
0.67 |
0.6 |
0.74 |
0.98 |
0.98 |
0.8 |
0.69 |
0.66 |
0.57 |
0.74 |
0.67 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.98 |
0.99 |
0.97 |
0.99 |
0.99 |
0.99 |
0.99 |
0.77 |
0.99 |
0.59 |
0.99 |
0.98 |
sipu/s2 |
0.92 |
0.91 |
0.81 |
0.95 |
0.92 |
0.92 |
0.8 |
0.78 |
0.94 |
0.26 |
0.94 |
0.91 |
sipu/s3 |
0.64 |
0.7 |
0.55 |
0.75 |
0.71 |
0.69 |
0.61 |
0.64 |
0.74 |
0.26 |
0.73 |
0.7 |
sipu/s4 |
0.55 |
0.59 |
0.49 |
0.67 |
0.64 |
0.62 |
0.53 |
0.58 |
0.66 |
0.26 |
0.61 |
0.58 |
sipu/spiral |
0.36 |
0.34 |
0.34 |
0.33 |
1 |
1 |
1 |
0.82 |
0.33 |
1 |
0.33 |
0.34 |
sipu/unbalance |
1 |
1 |
0.77 |
1 |
0.69 |
0.73 |
0.84 |
0.66 |
1 |
1 |
1 |
1 |
uci/ecoli |
0.82 |
0.61 |
0.72 |
0.71 |
0.51 |
0.59 |
0.75 |
0.48 |
0.59 |
0.53 |
0.62 |
0.61 |
uci/glass |
0.49 |
0.5 |
0.55 |
0.47 |
0.33 |
0.48 |
0.48 |
0.41 |
0.51 |
0.51 |
0.47 |
0.51 |
uci/ionosphere |
0.73 |
0.61 |
0.73 |
0.71 |
0.64 |
0.64 |
0.64 |
0.56 |
0.61 |
0.73 |
0.73 |
0.61 |
uci/sonar |
0.65 |
0.53 |
0.53 |
0.51 |
0.52 |
0.52 |
0.64 |
0.51 |
0.5 |
0.7 |
0.7 |
0.53 |
uci/statlog |
0.37 |
0.47 |
0.43 |
0.57 |
0.68 |
0.6 |
0.58 |
0.6 |
0.48 |
0.38 |
nan |
0.45 |
uci/wdbc |
0.72 |
0.74 |
0.72 |
0.87 |
0.6 |
0.74 |
0.74 |
0.82 |
0.79 |
0.73 |
0.73 |
0.74 |
uci/wine |
0.62 |
0.58 |
0.59 |
0.88 |
0.58 |
0.58 |
0.59 |
0.6 |
0.58 |
0.56 |
0.63 |
0.58 |
uci/yeast |
0.46 |
0.28 |
0.42 |
0.26 |
0.25 |
0.39 |
0.39 |
0.23 |
0.3 |
0.47 |
0.47 |
0.29 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.49 |
0.45 |
0.6 |
0.64 |
0.82 |
0.62 |
0.44 |
0.67 |
0.44 |
0.5 |
0.5 |
0.44 |
wut/graph |
0.49 |
0.51 |
0.48 |
0.88 |
0.54 |
0.55 |
0.49 |
0.57 |
0.51 |
0.31 |
0.49 |
0.53 |
wut/isolation |
0.34 |
0.35 |
0.34 |
0.33 |
1 |
1 |
1 |
1 |
0.33 |
1 |
0.33 |
0.34 |
wut/labirynth |
0.51 |
0.61 |
0.46 |
0.72 |
0.62 |
0.69 |
0.79 |
0.79 |
0.45 |
0.85 |
0.53 |
0.49 |
wut/mk1 |
0.99 |
0.99 |
0.98 |
0.99 |
0.99 |
0.99 |
0.99 |
0.7 |
0.99 |
0.77 |
0.99 |
0.99 |
wut/mk2 |
0.5 |
0.51 |
0.5 |
0.5 |
1 |
1 |
1 |
1 |
0.5 |
1 |
0.5 |
0.51 |
wut/mk3 |
0.77 |
0.9 |
0.9 |
0.92 |
0.86 |
0.86 |
0.75 |
0.7 |
0.93 |
0.57 |
0.92 |
0.91 |
wut/mk4 |
0.49 |
0.49 |
0.55 |
0.67 |
1 |
1 |
1 |
0.73 |
0.5 |
1 |
0.51 |
0.5 |
wut/olympic |
0.35 |
0.33 |
0.3 |
0.32 |
0.37 |
0.37 |
0.36 |
0.38 |
0.29 |
0.45 |
0.33 |
0.31 |
wut/smile |
0.99 |
0.73 |
0.86 |
0.73 |
0.75 |
1 |
1 |
0.72 |
0.73 |
1 |
1 |
0.76 |
wut/stripes |
0.51 |
0.54 |
0.53 |
0.51 |
1 |
1 |
1 |
1 |
0.51 |
1 |
0.51 |
0.55 |
wut/trajectories |
1 |
1 |
0.8 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.5 |
0.5 |
0.54 |
0.5 |
0.64 |
1 |
1 |
0.64 |
0.5 |
1 |
0.86 |
0.5 |
wut/twosplashes |
0.69 |
0.64 |
0.61 |
0.84 |
0.75 |
0.75 |
0.75 |
0.87 |
0.64 |
0.7 |
0.64 |
0.6 |
wut/windows |
0.39 |
0.39 |
0.39 |
0.41 |
0.43 |
0.5 |
1 |
0.58 |
0.37 |
1 |
0.43 |
0.4 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.73 |
0.98 |
1 |
0.8 |
0.87 |
0.87 |
0.72 |
0.79 |
0.98 |
0.57 |
0.6 |
0.98 |
wut/x3 |
0.97 |
0.99 |
0.62 |
0.96 |
0.91 |
0.97 |
0.97 |
0.76 |
1 |
0.66 |
0.69 |
0.99 |
wut/z1 |
0.51 |
0.42 |
0.46 |
0.36 |
0.6 |
0.6 |
0.5 |
0.6 |
0.47 |
0.55 |
0.43 |
0.42 |
wut/z2 |
0.68 |
0.66 |
0.62 |
1 |
0.66 |
0.76 |
0.89 |
0.65 |
0.64 |
0.86 |
0.99 |
0.61 |
wut/z3 |
1 |
1 |
0.95 |
1 |
0.74 |
0.77 |
0.94 |
0.74 |
1 |
0.84 |
0.99 |
1 |
ami¶
Summary statistics for ami (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.57 |
0.36 |
0 |
0.27 |
0.65 |
0.95 |
1 |
Birch_0.01 |
73 |
0.61 |
0.33 |
0 |
0.34 |
0.68 |
0.95 |
1 |
Complete linkage |
73 |
0.56 |
0.32 |
0 |
0.35 |
0.56 |
0.88 |
1 |
Gaussian mixtures |
73 |
0.68 |
0.34 |
0 |
0.43 |
0.8 |
0.98 |
1 |
Genie_0.1 |
73 |
0.79 |
0.26 |
0 |
0.71 |
0.87 |
1 |
1 |
Genie_0.3 |
73 |
0.82 |
0.24 |
0 |
0.74 |
0.95 |
1 |
1 |
Genie_0.5 |
73 |
0.82 |
0.25 |
0.06 |
0.74 |
0.94 |
1 |
1 |
ITM |
73 |
0.75 |
0.24 |
0 |
0.63 |
0.77 |
0.97 |
1 |
K-means |
73 |
0.6 |
0.33 |
0 |
0.36 |
0.65 |
0.97 |
1 |
Single linkage |
73 |
0.49 |
0.46 |
0 |
0 |
0.71 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.67 |
0.34 |
0 |
0.47 |
0.76 |
0.99 |
1 |
Ward linkage |
73 |
0.61 |
0.32 |
0 |
0.35 |
0.65 |
0.93 |
1 |
Ranks for ami (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.5 |
3.6 |
1 |
3.8 |
6 |
9 |
12 |
Birch_0.01 |
72 |
5.7 |
3 |
1 |
3.8 |
6 |
8 |
12 |
Complete linkage |
72 |
7.6 |
3.3 |
1 |
6 |
8 |
11 |
12 |
Gaussian mixtures |
72 |
4.4 |
3.7 |
1 |
1 |
3 |
7 |
12 |
Genie_0.1 |
72 |
3.7 |
3.2 |
1 |
1 |
3 |
5 |
12 |
Genie_0.3 |
72 |
3.1 |
2.9 |
1 |
1 |
1.5 |
4.2 |
11 |
Genie_0.5 |
72 |
3.9 |
3.6 |
1 |
1 |
1.5 |
7.2 |
12 |
ITM |
72 |
5.6 |
3.9 |
1 |
1.8 |
5 |
9 |
12 |
K-means |
72 |
5.6 |
3.9 |
1 |
1 |
6 |
9 |
12 |
Single linkage |
72 |
7.5 |
5 |
1 |
1 |
11 |
12 |
12 |
Spectral_RBF_5 |
72 |
5 |
3.5 |
1 |
1 |
4.5 |
8 |
11 |
Ward linkage |
72 |
5.9 |
3.1 |
1 |
3.8 |
6 |
8 |
12 |
Raw results for ami (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.22 |
0.22 |
0.2 |
0.13 |
1 |
1 |
1 |
1 |
0.29 |
1 |
1 |
0.22 |
fcps/chainlink |
0.36 |
0.37 |
0.39 |
0.84 |
1 |
1 |
1 |
1 |
0.07 |
1 |
1 |
0.37 |
fcps/engytime |
0.16 |
0.68 |
0.15 |
0.96 |
0.79 |
0.79 |
0.79 |
0.74 |
0.77 |
0 |
0.87 |
0.68 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.94 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.5 |
0.53 |
0.53 |
1 |
1 |
1 |
1 |
1 |
0.54 |
1 |
0.82 |
0.51 |
fcps/target |
0.33 |
0.64 |
0.38 |
0.65 |
1 |
1 |
1 |
1 |
0.63 |
1 |
1 |
0.63 |
fcps/tetra |
0.99 |
0.96 |
0.98 |
1 |
1 |
1 |
1 |
1 |
1 |
0 |
1 |
0.96 |
fcps/twodiamonds |
0.99 |
0.99 |
0.97 |
1 |
0.95 |
0.95 |
0.95 |
0.97 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.78 |
1 |
1 |
1 |
1 |
0.77 |
1 |
0.68 |
0.49 |
graves/dense |
0.88 |
0.88 |
0.35 |
1 |
0.93 |
0.93 |
0.93 |
1 |
0.76 |
0.02 |
0.88 |
0.88 |
graves/fuzzyx |
0.8 |
0.84 |
0.9 |
0.99 |
0.79 |
0.64 |
0.59 |
0.94 |
0.99 |
0 |
0.69 |
0.75 |
graves/line |
0.14 |
0.14 |
0.14 |
1 |
0.2 |
1 |
1 |
0.25 |
0.18 |
1 |
1 |
0.1 |
graves/parabolic |
0.49 |
0.57 |
0.51 |
0.43 |
0.74 |
0.74 |
0.74 |
0.61 |
0.48 |
0 |
0.55 |
0.52 |
graves/ring |
0.23 |
0.24 |
0.31 |
0.13 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
0.24 |
graves/ring_noisy |
0 |
0.23 |
0.36 |
0.12 |
1 |
1 |
1 |
1 |
0.28 |
0 |
1 |
0.26 |
graves/ring_outliers |
0.65 |
0.65 |
0.41 |
0.65 |
1 |
1 |
1 |
1 |
0.65 |
1 |
1 |
0.65 |
graves/zigzag |
0.67 |
0.76 |
0.56 |
0.96 |
1 |
1 |
1 |
0.85 |
0.71 |
1 |
0.8 |
0.71 |
graves/zigzag_noisy |
0.66 |
0.75 |
0.67 |
0.89 |
0.85 |
1 |
1 |
0.68 |
0.66 |
0.74 |
0.78 |
0.76 |
graves/zigzag_outliers |
0.49 |
0.65 |
0.53 |
0.97 |
1 |
1 |
1 |
0.9 |
0.61 |
0.74 |
0.53 |
0.57 |
other/chameleon_t4_8k |
0.76 |
0.73 |
0.62 |
0.69 |
0.91 |
1 |
0.95 |
0.87 |
0.7 |
0 |
0.72 |
0.73 |
other/chameleon_t5_8k |
1 |
1 |
0.83 |
1 |
1 |
1 |
0.93 |
0.75 |
1 |
0 |
1 |
1 |
other/chameleon_t7_10k |
0.69 |
0.71 |
0.6 |
0.68 |
0.78 |
0.87 |
1 |
0.76 |
0.66 |
0 |
0.64 |
0.69 |
other/chameleon_t8_8k |
0.59 |
0.58 |
0.55 |
0.64 |
0.79 |
0.79 |
0.86 |
0.76 |
0.59 |
0 |
0.58 |
0.59 |
other/hdbscan |
0.62 |
0.75 |
0.61 |
0.82 |
0.97 |
0.87 |
0.87 |
0.85 |
0.73 |
0 |
0.55 |
0.86 |
other/iris |
0.8 |
0.77 |
0.72 |
0.9 |
0.87 |
0.87 |
0.7 |
0.87 |
0.76 |
0.71 |
0.8 |
0.77 |
other/iris5 |
0.63 |
0.56 |
0.46 |
0.81 |
0.58 |
0.58 |
0.76 |
0.54 |
0.54 |
0.34 |
0.61 |
0.56 |
other/square |
0.27 |
0 |
0.28 |
0.03 |
1 |
1 |
1 |
1 |
0.02 |
1 |
0.28 |
0.35 |
sipu/a1 |
0.95 |
0.96 |
0.95 |
0.97 |
0.96 |
0.95 |
0.94 |
0.89 |
0.97 |
0.78 |
0.96 |
0.95 |
sipu/a2 |
0.96 |
0.96 |
0.95 |
0.98 |
0.97 |
0.96 |
0.94 |
0.9 |
0.98 |
0.76 |
0.96 |
0.96 |
sipu/a3 |
0.97 |
0.97 |
0.96 |
0.97 |
0.97 |
0.97 |
0.95 |
0.91 |
0.97 |
0.76 |
0.96 |
0.97 |
sipu/aggregation |
1 |
0.92 |
0.9 |
1 |
0.7 |
0.76 |
0.92 |
0.78 |
0.88 |
0.88 |
0.99 |
0.92 |
sipu/compound |
0.93 |
0.88 |
0.93 |
0.93 |
0.85 |
0.85 |
0.88 |
0.74 |
0.83 |
0.93 |
0.86 |
0.88 |
sipu/d31 |
0.95 |
0.95 |
0.95 |
0.96 |
0.96 |
0.95 |
0.9 |
0.91 |
0.97 |
0.63 |
0.96 |
0.95 |
sipu/flame |
0.48 |
0.35 |
0.12 |
0.42 |
1 |
1 |
1 |
0.43 |
0.43 |
0.02 |
0.73 |
0.35 |
sipu/jain |
0.7 |
0.5 |
0.7 |
0.2 |
0.23 |
1 |
1 |
0.39 |
0.37 |
0.24 |
0.5 |
0.5 |
sipu/pathbased |
0.64 |
0.59 |
0.5 |
0.66 |
0.95 |
0.95 |
0.81 |
0.61 |
0.58 |
0 |
0.67 |
0.59 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.98 |
0.99 |
0.98 |
0.99 |
0.99 |
0.99 |
0.99 |
0.88 |
0.99 |
0.79 |
0.99 |
0.98 |
sipu/s2 |
0.93 |
0.92 |
0.88 |
0.95 |
0.93 |
0.93 |
0.91 |
0.86 |
0.95 |
0 |
0.94 |
0.93 |
sipu/s3 |
0.75 |
0.77 |
0.7 |
0.8 |
0.78 |
0.77 |
0.75 |
0.74 |
0.79 |
0 |
0.79 |
0.77 |
sipu/s4 |
0.66 |
0.69 |
0.63 |
0.73 |
0.71 |
0.7 |
0.66 |
0.68 |
0.72 |
0 |
0.71 |
0.69 |
sipu/spiral |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
0.78 |
0 |
1 |
0 |
0 |
sipu/unbalance |
1 |
1 |
0.82 |
1 |
0.75 |
0.77 |
0.82 |
0.75 |
1 |
0.99 |
1 |
1 |
uci/ecoli |
0.71 |
0.62 |
0.64 |
0.57 |
0.49 |
0.54 |
0.57 |
0.49 |
0.58 |
0.11 |
0.48 |
0.62 |
uci/glass |
0.07 |
0.34 |
0.35 |
0.33 |
0.25 |
0.38 |
0.34 |
0.33 |
0.4 |
0.03 |
0.31 |
0.37 |
uci/ionosphere |
0 |
0.14 |
0.01 |
0.32 |
0.13 |
0.13 |
0.06 |
0.09 |
0.13 |
0 |
0 |
0.14 |
uci/sonar |
0 |
0 |
0 |
0 |
0 |
0 |
0.07 |
0 |
0.01 |
0 |
0 |
0 |
uci/statlog |
0.01 |
0.53 |
0.35 |
0.61 |
0.68 |
0.68 |
0.7 |
0.63 |
0.52 |
0 |
nan |
0.49 |
uci/wdbc |
0.09 |
0.32 |
0.09 |
0.61 |
0.24 |
0.31 |
0.31 |
0.51 |
0.46 |
0 |
0 |
0.32 |
uci/wine |
0.4 |
0.41 |
0.44 |
0.82 |
0.41 |
0.41 |
0.38 |
0.37 |
0.42 |
0.04 |
0.42 |
0.41 |
uci/yeast |
0.05 |
0.22 |
0.18 |
0.14 |
0.22 |
0.25 |
0.19 |
0.19 |
0.26 |
0.05 |
0.06 |
0.23 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.04 |
0.18 |
0.48 |
0.61 |
0.81 |
0.62 |
0.36 |
0.7 |
0.36 |
0 |
0.01 |
0.31 |
wut/graph |
0.62 |
0.63 |
0.6 |
0.89 |
0.68 |
0.69 |
0.62 |
0.69 |
0.64 |
0.23 |
0.62 |
0.65 |
wut/isolation |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
wut/labirynth |
0.59 |
0.71 |
0.54 |
0.76 |
0.71 |
0.78 |
0.88 |
0.87 |
0.5 |
0.85 |
0.6 |
0.58 |
wut/mk1 |
0.98 |
0.98 |
0.95 |
0.98 |
0.98 |
0.98 |
0.98 |
0.61 |
0.98 |
0.72 |
0.98 |
0.98 |
wut/mk2 |
0 |
0.01 |
0.01 |
0 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0 |
0 |
wut/mk3 |
0.71 |
0.83 |
0.83 |
0.85 |
0.8 |
0.8 |
0.69 |
0.61 |
0.86 |
0 |
0.85 |
0.84 |
wut/mk4 |
0.11 |
0.2 |
0.3 |
0.58 |
1 |
1 |
1 |
0.65 |
0.25 |
1 |
0.25 |
0.24 |
wut/olympic |
0.31 |
0.27 |
0.21 |
0.25 |
0.33 |
0.33 |
0.31 |
0.31 |
0.2 |
0 |
0.31 |
0.23 |
wut/smile |
0.98 |
0.79 |
0.83 |
0.79 |
0.8 |
1 |
1 |
0.85 |
0.79 |
1 |
1 |
0.8 |
wut/stripes |
0 |
0 |
0.01 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.01 |
wut/trajectories |
1 |
1 |
0.83 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.35 |
0.36 |
0.45 |
0.36 |
0.62 |
1 |
1 |
0.62 |
0.38 |
1 |
0.74 |
0.39 |
wut/twosplashes |
0.04 |
0.21 |
0.17 |
0.56 |
0.4 |
0.4 |
0.4 |
0.69 |
0.21 |
0.01 |
0.21 |
0.15 |
wut/windows |
0.4 |
0.4 |
0.4 |
0.43 |
0.48 |
0.56 |
1 |
0.65 |
0.35 |
1 |
0.43 |
0.4 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.72 |
0.97 |
1 |
0.75 |
0.84 |
0.84 |
0.72 |
0.77 |
0.97 |
0 |
0.41 |
0.97 |
wut/x3 |
0.91 |
0.97 |
0.63 |
0.91 |
0.87 |
0.91 |
0.91 |
0.69 |
1 |
0.01 |
0.68 |
0.97 |
wut/z1 |
0.32 |
0.13 |
0.26 |
0.03 |
0.47 |
0.47 |
0.24 |
0.46 |
0.27 |
0.05 |
0.19 |
0.13 |
wut/z2 |
0.72 |
0.72 |
0.64 |
1 |
0.72 |
0.74 |
0.86 |
0.71 |
0.68 |
0.81 |
0.98 |
0.69 |
wut/z3 |
0.99 |
0.99 |
0.93 |
0.99 |
0.74 |
0.74 |
0.91 |
0.75 |
1 |
0.84 |
0.97 |
1 |
nmi¶
Summary statistics for nmi (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.58 |
0.36 |
0 |
0.27 |
0.66 |
0.95 |
1 |
Birch_0.01 |
73 |
0.61 |
0.33 |
0 |
0.35 |
0.68 |
0.96 |
1 |
Complete linkage |
73 |
0.57 |
0.32 |
0 |
0.35 |
0.57 |
0.88 |
1 |
Gaussian mixtures |
73 |
0.68 |
0.34 |
0 |
0.43 |
0.8 |
0.98 |
1 |
Genie_0.1 |
73 |
0.79 |
0.26 |
0 |
0.71 |
0.87 |
1 |
1 |
Genie_0.3 |
73 |
0.83 |
0.24 |
0 |
0.74 |
0.95 |
1 |
1 |
Genie_0.5 |
73 |
0.82 |
0.25 |
0.07 |
0.74 |
0.94 |
1 |
1 |
ITM |
73 |
0.75 |
0.24 |
0.01 |
0.63 |
0.78 |
0.97 |
1 |
K-means |
73 |
0.61 |
0.33 |
0 |
0.37 |
0.65 |
0.97 |
1 |
Single linkage |
73 |
0.5 |
0.45 |
0 |
0.01 |
0.72 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.67 |
0.34 |
0 |
0.49 |
0.76 |
0.99 |
1 |
Ward linkage |
73 |
0.61 |
0.32 |
0 |
0.35 |
0.66 |
0.93 |
1 |
Ranks for nmi (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.6 |
3.6 |
1 |
4 |
6.5 |
9.2 |
12 |
Birch_0.01 |
72 |
5.8 |
2.9 |
1 |
4 |
6 |
8 |
12 |
Complete linkage |
72 |
7.7 |
3.3 |
1 |
6 |
8.5 |
11 |
12 |
Gaussian mixtures |
72 |
4.4 |
3.7 |
1 |
1 |
3 |
7.2 |
12 |
Genie_0.1 |
72 |
3.8 |
3.3 |
1 |
1 |
2.5 |
5.2 |
12 |
Genie_0.3 |
72 |
3.1 |
2.9 |
1 |
1 |
1.5 |
4 |
11 |
Genie_0.5 |
72 |
3.8 |
3.6 |
1 |
1 |
1.5 |
7.2 |
12 |
ITM |
72 |
5.5 |
3.9 |
1 |
1 |
5.5 |
9 |
12 |
K-means |
72 |
5.7 |
3.9 |
1 |
1 |
6 |
9 |
12 |
Single linkage |
72 |
7.5 |
5 |
1 |
1 |
10.5 |
12 |
12 |
Spectral_RBF_5 |
72 |
5 |
3.5 |
1 |
1 |
4 |
8 |
11 |
Ward linkage |
72 |
6 |
3.1 |
1 |
4 |
6 |
8 |
12 |
Raw results for nmi (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.22 |
0.22 |
0.2 |
0.13 |
1 |
1 |
1 |
1 |
0.29 |
1 |
1 |
0.22 |
fcps/chainlink |
0.36 |
0.37 |
0.39 |
0.84 |
1 |
1 |
1 |
1 |
0.07 |
1 |
1 |
0.37 |
fcps/engytime |
0.16 |
0.68 |
0.15 |
0.96 |
0.79 |
0.79 |
0.79 |
0.74 |
0.77 |
0 |
0.87 |
0.68 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.95 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.5 |
0.53 |
0.53 |
1 |
1 |
1 |
1 |
1 |
0.54 |
1 |
0.82 |
0.51 |
fcps/target |
0.34 |
0.64 |
0.38 |
0.65 |
1 |
1 |
1 |
1 |
0.64 |
1 |
1 |
0.64 |
fcps/tetra |
0.99 |
0.96 |
0.98 |
1 |
1 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.96 |
fcps/twodiamonds |
0.99 |
0.99 |
0.97 |
1 |
0.95 |
0.95 |
0.95 |
0.97 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.78 |
1 |
1 |
1 |
1 |
0.77 |
1 |
0.68 |
0.49 |
graves/dense |
0.88 |
0.88 |
0.35 |
1 |
0.93 |
0.93 |
0.93 |
1 |
0.76 |
0.03 |
0.88 |
0.88 |
graves/fuzzyx |
0.8 |
0.84 |
0.9 |
0.99 |
0.79 |
0.64 |
0.59 |
0.94 |
0.99 |
0.01 |
0.69 |
0.75 |
graves/line |
0.14 |
0.14 |
0.14 |
1 |
0.2 |
1 |
1 |
0.25 |
0.18 |
1 |
1 |
0.1 |
graves/parabolic |
0.49 |
0.57 |
0.51 |
0.43 |
0.74 |
0.74 |
0.74 |
0.61 |
0.48 |
0.01 |
0.55 |
0.52 |
graves/ring |
0.23 |
0.24 |
0.31 |
0.13 |
1 |
1 |
1 |
1 |
0 |
1 |
1 |
0.24 |
graves/ring_noisy |
0 |
0.24 |
0.36 |
0.12 |
1 |
1 |
1 |
1 |
0.28 |
0 |
1 |
0.27 |
graves/ring_outliers |
0.66 |
0.66 |
0.41 |
0.65 |
1 |
1 |
1 |
1 |
0.65 |
1 |
1 |
0.66 |
graves/zigzag |
0.67 |
0.76 |
0.57 |
0.96 |
1 |
1 |
1 |
0.85 |
0.71 |
1 |
0.81 |
0.71 |
graves/zigzag_noisy |
0.67 |
0.76 |
0.68 |
0.89 |
0.85 |
1 |
1 |
0.68 |
0.67 |
0.74 |
0.79 |
0.77 |
graves/zigzag_outliers |
0.5 |
0.66 |
0.54 |
0.98 |
1 |
1 |
1 |
0.9 |
0.62 |
0.74 |
0.53 |
0.57 |
other/chameleon_t4_8k |
0.76 |
0.73 |
0.62 |
0.69 |
0.91 |
1 |
0.95 |
0.87 |
0.7 |
0 |
0.72 |
0.73 |
other/chameleon_t5_8k |
1 |
1 |
0.83 |
1 |
1 |
1 |
0.93 |
0.75 |
1 |
0 |
1 |
1 |
other/chameleon_t7_10k |
0.69 |
0.71 |
0.6 |
0.68 |
0.78 |
0.87 |
1 |
0.76 |
0.66 |
0 |
0.64 |
0.69 |
other/chameleon_t8_8k |
0.59 |
0.58 |
0.55 |
0.64 |
0.79 |
0.79 |
0.86 |
0.76 |
0.59 |
0 |
0.58 |
0.59 |
other/hdbscan |
0.62 |
0.75 |
0.61 |
0.82 |
0.97 |
0.87 |
0.88 |
0.85 |
0.73 |
0 |
0.55 |
0.86 |
other/iris |
0.81 |
0.77 |
0.72 |
0.9 |
0.87 |
0.87 |
0.71 |
0.87 |
0.76 |
0.72 |
0.8 |
0.77 |
other/iris5 |
0.64 |
0.57 |
0.47 |
0.81 |
0.59 |
0.59 |
0.76 |
0.55 |
0.55 |
0.36 |
0.62 |
0.57 |
other/square |
0.27 |
0 |
0.28 |
0.03 |
1 |
1 |
1 |
1 |
0.02 |
1 |
0.28 |
0.35 |
sipu/a1 |
0.95 |
0.96 |
0.95 |
0.97 |
0.96 |
0.95 |
0.94 |
0.89 |
0.97 |
0.79 |
0.96 |
0.95 |
sipu/a2 |
0.96 |
0.96 |
0.95 |
0.98 |
0.97 |
0.96 |
0.95 |
0.91 |
0.98 |
0.77 |
0.96 |
0.96 |
sipu/a3 |
0.97 |
0.97 |
0.96 |
0.98 |
0.98 |
0.97 |
0.95 |
0.91 |
0.98 |
0.76 |
0.97 |
0.97 |
sipu/aggregation |
1 |
0.92 |
0.9 |
1 |
0.71 |
0.76 |
0.92 |
0.78 |
0.88 |
0.88 |
0.99 |
0.92 |
sipu/compound |
0.93 |
0.88 |
0.93 |
0.93 |
0.85 |
0.85 |
0.89 |
0.75 |
0.83 |
0.93 |
0.86 |
0.88 |
sipu/d31 |
0.95 |
0.96 |
0.95 |
0.96 |
0.96 |
0.95 |
0.91 |
0.91 |
0.97 |
0.64 |
0.96 |
0.95 |
sipu/flame |
0.48 |
0.35 |
0.13 |
0.42 |
1 |
1 |
1 |
0.43 |
0.43 |
0.02 |
0.73 |
0.35 |
sipu/jain |
0.7 |
0.51 |
0.7 |
0.2 |
0.23 |
1 |
1 |
0.39 |
0.37 |
0.25 |
0.51 |
0.51 |
sipu/pathbased |
0.64 |
0.6 |
0.51 |
0.66 |
0.95 |
0.95 |
0.81 |
0.61 |
0.59 |
0.02 |
0.67 |
0.6 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.98 |
0.99 |
0.98 |
0.99 |
0.99 |
0.99 |
0.99 |
0.88 |
0.99 |
0.79 |
0.99 |
0.98 |
sipu/s2 |
0.93 |
0.92 |
0.88 |
0.95 |
0.93 |
0.93 |
0.91 |
0.86 |
0.95 |
0.01 |
0.94 |
0.93 |
sipu/s3 |
0.75 |
0.77 |
0.71 |
0.8 |
0.78 |
0.78 |
0.75 |
0.75 |
0.79 |
0.01 |
0.79 |
0.77 |
sipu/s4 |
0.67 |
0.69 |
0.63 |
0.73 |
0.72 |
0.71 |
0.67 |
0.68 |
0.72 |
0.01 |
0.71 |
0.69 |
sipu/spiral |
0 |
0.01 |
0.01 |
0 |
1 |
1 |
1 |
0.79 |
0 |
1 |
0 |
0.01 |
sipu/unbalance |
1 |
1 |
0.82 |
1 |
0.75 |
0.77 |
0.82 |
0.75 |
1 |
0.99 |
1 |
1 |
uci/ecoli |
0.72 |
0.63 |
0.65 |
0.59 |
0.51 |
0.56 |
0.59 |
0.51 |
0.6 |
0.15 |
0.51 |
0.63 |
uci/glass |
0.11 |
0.37 |
0.38 |
0.36 |
0.28 |
0.41 |
0.37 |
0.35 |
0.43 |
0.07 |
0.34 |
0.39 |
uci/ionosphere |
0.01 |
0.14 |
0.02 |
0.32 |
0.13 |
0.13 |
0.07 |
0.09 |
0.13 |
0.01 |
0.01 |
0.14 |
uci/sonar |
0.01 |
0 |
0 |
0.01 |
0 |
0 |
0.08 |
0.01 |
0.01 |
0.01 |
0.01 |
0 |
uci/statlog |
0.02 |
0.53 |
0.35 |
0.62 |
0.68 |
0.68 |
0.7 |
0.63 |
0.52 |
0.01 |
nan |
0.49 |
uci/wdbc |
0.09 |
0.32 |
0.09 |
0.61 |
0.24 |
0.32 |
0.32 |
0.51 |
0.46 |
0.01 |
0.01 |
0.32 |
uci/wine |
0.4 |
0.42 |
0.44 |
0.82 |
0.42 |
0.42 |
0.39 |
0.38 |
0.43 |
0.06 |
0.43 |
0.42 |
uci/yeast |
0.07 |
0.23 |
0.19 |
0.15 |
0.23 |
0.27 |
0.2 |
0.2 |
0.27 |
0.07 |
0.07 |
0.24 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.04 |
0.19 |
0.48 |
0.61 |
0.81 |
0.62 |
0.36 |
0.7 |
0.37 |
0 |
0.01 |
0.31 |
wut/graph |
0.62 |
0.63 |
0.61 |
0.89 |
0.68 |
0.69 |
0.62 |
0.7 |
0.64 |
0.24 |
0.63 |
0.66 |
wut/isolation |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
0 |
1 |
0 |
0 |
wut/labirynth |
0.59 |
0.71 |
0.54 |
0.76 |
0.71 |
0.78 |
0.88 |
0.87 |
0.5 |
0.85 |
0.6 |
0.58 |
wut/mk1 |
0.98 |
0.98 |
0.95 |
0.98 |
0.98 |
0.98 |
0.98 |
0.62 |
0.98 |
0.73 |
0.98 |
0.98 |
wut/mk2 |
0 |
0.01 |
0.01 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0 |
0 |
wut/mk3 |
0.71 |
0.83 |
0.83 |
0.85 |
0.8 |
0.8 |
0.69 |
0.61 |
0.86 |
0.01 |
0.85 |
0.84 |
wut/mk4 |
0.12 |
0.2 |
0.3 |
0.58 |
1 |
1 |
1 |
0.65 |
0.25 |
1 |
0.25 |
0.24 |
wut/olympic |
0.31 |
0.27 |
0.21 |
0.25 |
0.33 |
0.34 |
0.31 |
0.31 |
0.2 |
0 |
0.31 |
0.23 |
wut/smile |
0.98 |
0.79 |
0.83 |
0.79 |
0.8 |
1 |
1 |
0.85 |
0.79 |
1 |
1 |
0.81 |
wut/stripes |
0 |
0 |
0.01 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.01 |
wut/trajectories |
1 |
1 |
0.83 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.35 |
0.36 |
0.45 |
0.36 |
0.63 |
1 |
1 |
0.62 |
0.38 |
1 |
0.74 |
0.39 |
wut/twosplashes |
0.04 |
0.21 |
0.17 |
0.56 |
0.4 |
0.4 |
0.4 |
0.69 |
0.21 |
0.01 |
0.21 |
0.15 |
wut/windows |
0.41 |
0.41 |
0.4 |
0.43 |
0.48 |
0.56 |
1 |
0.65 |
0.35 |
1 |
0.43 |
0.4 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.73 |
0.97 |
1 |
0.75 |
0.85 |
0.85 |
0.73 |
0.78 |
0.97 |
0.03 |
0.43 |
0.97 |
wut/x3 |
0.91 |
0.97 |
0.64 |
0.91 |
0.87 |
0.91 |
0.91 |
0.7 |
1 |
0.04 |
0.69 |
0.97 |
wut/z1 |
0.33 |
0.14 |
0.27 |
0.04 |
0.47 |
0.47 |
0.24 |
0.47 |
0.27 |
0.07 |
0.19 |
0.14 |
wut/z2 |
0.72 |
0.72 |
0.64 |
1 |
0.72 |
0.74 |
0.86 |
0.72 |
0.68 |
0.81 |
0.98 |
0.69 |
wut/z3 |
0.99 |
0.99 |
0.93 |
0.99 |
0.74 |
0.75 |
0.91 |
0.75 |
1 |
0.84 |
0.97 |
1 |
npa¶
Summary statistics for npa (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.59 |
0.34 |
0 |
0.32 |
0.6 |
0.96 |
1 |
Birch_0.01 |
73 |
0.64 |
0.31 |
0.01 |
0.44 |
0.67 |
0.96 |
1 |
Complete linkage |
73 |
0.59 |
0.3 |
0.02 |
0.39 |
0.56 |
0.89 |
1 |
Gaussian mixtures |
73 |
0.71 |
0.33 |
0.01 |
0.51 |
0.86 |
0.99 |
1 |
Genie_0.1 |
73 |
0.79 |
0.26 |
0.02 |
0.63 |
0.94 |
1 |
1 |
Genie_0.3 |
73 |
0.83 |
0.22 |
0.02 |
0.68 |
0.96 |
1 |
1 |
Genie_0.5 |
73 |
0.81 |
0.25 |
0.08 |
0.71 |
0.92 |
1 |
1 |
ITM |
73 |
0.75 |
0.23 |
0.08 |
0.6 |
0.8 |
0.99 |
1 |
K-means |
73 |
0.64 |
0.31 |
0.01 |
0.42 |
0.65 |
0.97 |
1 |
Single linkage |
73 |
0.49 |
0.42 |
0 |
0.07 |
0.37 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.7 |
0.32 |
0.01 |
0.42 |
0.83 |
1 |
1 |
Ward linkage |
73 |
0.64 |
0.29 |
0.02 |
0.41 |
0.63 |
0.95 |
1 |
Ranks for npa (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.3 |
3.8 |
1 |
2.8 |
7 |
9.2 |
12 |
Birch_0.01 |
72 |
5.9 |
3.1 |
1 |
4 |
6 |
8 |
12 |
Complete linkage |
72 |
7.6 |
3.1 |
1 |
6.8 |
8 |
10 |
12 |
Gaussian mixtures |
72 |
4.3 |
3.9 |
1 |
1 |
3 |
7 |
12 |
Genie_0.1 |
72 |
4 |
3.6 |
1 |
1 |
2 |
7 |
12 |
Genie_0.3 |
72 |
3.5 |
3.2 |
1 |
1 |
2 |
6 |
11 |
Genie_0.5 |
72 |
4.1 |
3.8 |
1 |
1 |
1.5 |
7.2 |
12 |
ITM |
72 |
5.4 |
3.9 |
1 |
1 |
5 |
9 |
12 |
K-means |
72 |
5.7 |
3.8 |
1 |
1 |
6.5 |
9 |
12 |
Single linkage |
72 |
7.5 |
5.1 |
1 |
1 |
11 |
12 |
12 |
Spectral_RBF_5 |
72 |
5.1 |
3.6 |
1 |
1 |
5 |
8 |
12 |
Ward linkage |
72 |
6.1 |
3.2 |
1 |
4 |
6 |
8 |
12 |
Raw results for npa (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.31 |
0.31 |
0.29 |
0.17 |
1 |
1 |
1 |
1 |
0.43 |
1 |
1 |
0.31 |
fcps/chainlink |
0.52 |
0.53 |
0.56 |
0.95 |
1 |
1 |
1 |
1 |
0.31 |
1 |
1 |
0.53 |
fcps/engytime |
0.23 |
0.88 |
0.2 |
0.99 |
0.92 |
0.92 |
0.92 |
0.91 |
0.92 |
0 |
0.96 |
0.86 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.94 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.58 |
0.62 |
0.62 |
1 |
1 |
1 |
1 |
1 |
0.65 |
1 |
0.9 |
0.58 |
fcps/target |
0.6 |
0.6 |
0.66 |
0.58 |
1 |
1 |
1 |
1 |
0.55 |
1 |
1 |
0.59 |
fcps/tetra |
1 |
0.98 |
0.99 |
1 |
1 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.98 |
fcps/twodiamonds |
1 |
1 |
0.99 |
1 |
0.99 |
0.99 |
0.99 |
0.99 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.93 |
1 |
1 |
1 |
1 |
0.93 |
1 |
0.88 |
0.78 |
graves/dense |
0.96 |
0.96 |
0.51 |
1 |
0.98 |
0.98 |
0.98 |
1 |
0.9 |
0.03 |
0.96 |
0.96 |
graves/fuzzyx |
0.86 |
0.95 |
0.97 |
1 |
0.84 |
0.72 |
0.63 |
0.97 |
1 |
0.03 |
0.78 |
0.88 |
graves/line |
0.01 |
0.01 |
0.02 |
1 |
0.19 |
1 |
1 |
0.35 |
0.14 |
1 |
1 |
0.19 |
graves/parabolic |
0.77 |
0.76 |
0.77 |
0.73 |
0.9 |
0.9 |
0.9 |
0.8 |
0.77 |
0.02 |
0.81 |
0.79 |
graves/ring |
0.34 |
0.34 |
0.46 |
0.17 |
1 |
1 |
1 |
1 |
0.01 |
1 |
1 |
0.34 |
graves/ring_noisy |
0 |
0.34 |
0.52 |
0.16 |
1 |
1 |
1 |
1 |
0.4 |
0 |
1 |
0.38 |
graves/ring_outliers |
0.58 |
0.57 |
0.53 |
0.55 |
1 |
1 |
1 |
1 |
0.55 |
1 |
1 |
0.58 |
graves/zigzag |
0.69 |
0.73 |
0.56 |
0.98 |
1 |
1 |
1 |
0.86 |
0.7 |
1 |
0.82 |
0.63 |
graves/zigzag_noisy |
0.65 |
0.77 |
0.58 |
0.95 |
0.88 |
1 |
1 |
0.67 |
0.64 |
0.5 |
0.78 |
0.79 |
graves/zigzag_outliers |
0.34 |
0.53 |
0.46 |
0.99 |
1 |
1 |
1 |
0.9 |
0.51 |
0.53 |
0.4 |
0.42 |
other/chameleon_t4_8k |
0.72 |
0.63 |
0.6 |
0.63 |
0.77 |
1 |
0.89 |
0.83 |
0.63 |
0.09 |
0.64 |
0.62 |
other/chameleon_t5_8k |
1 |
1 |
0.79 |
1 |
1 |
1 |
0.82 |
0.69 |
1 |
0.01 |
1 |
1 |
other/chameleon_t7_10k |
0.49 |
0.45 |
0.43 |
0.4 |
0.55 |
0.77 |
1 |
0.61 |
0.44 |
0.21 |
0.41 |
0.51 |
other/chameleon_t8_8k |
0.41 |
0.46 |
0.39 |
0.51 |
0.6 |
0.6 |
0.79 |
0.58 |
0.4 |
0.09 |
0.4 |
0.4 |
other/hdbscan |
0.4 |
0.72 |
0.54 |
0.86 |
0.98 |
0.74 |
0.74 |
0.86 |
0.77 |
0.07 |
0.47 |
0.91 |
other/iris |
0.86 |
0.84 |
0.76 |
0.95 |
0.94 |
0.94 |
0.55 |
0.94 |
0.84 |
0.52 |
0.85 |
0.84 |
other/iris5 |
0.8 |
0.77 |
0.66 |
0.93 |
0.66 |
0.66 |
0.91 |
0.54 |
0.77 |
0.31 |
0.79 |
0.77 |
other/square |
0.39 |
0.01 |
0.41 |
0.19 |
1 |
1 |
1 |
1 |
0.17 |
1 |
0.41 |
0.5 |
sipu/a1 |
0.96 |
0.96 |
0.96 |
0.98 |
0.97 |
0.91 |
0.84 |
0.82 |
0.98 |
0.37 |
0.97 |
0.95 |
sipu/a2 |
0.97 |
0.97 |
0.95 |
0.98 |
0.97 |
0.94 |
0.83 |
0.83 |
0.98 |
0.3 |
0.93 |
0.96 |
sipu/a3 |
0.97 |
0.97 |
0.96 |
0.96 |
0.98 |
0.95 |
0.84 |
0.83 |
0.96 |
0.25 |
0.95 |
0.97 |
sipu/aggregation |
1 |
0.82 |
0.75 |
1 |
0.55 |
0.63 |
0.86 |
0.63 |
0.75 |
0.79 |
1 |
0.81 |
sipu/compound |
0.94 |
0.81 |
0.94 |
0.94 |
0.75 |
0.76 |
0.89 |
0.66 |
0.73 |
0.94 |
0.82 |
0.81 |
sipu/d31 |
0.94 |
0.96 |
0.96 |
0.97 |
0.97 |
0.93 |
0.76 |
0.85 |
0.98 |
0.24 |
0.97 |
0.96 |
sipu/flame |
0.67 |
0.47 |
0.03 |
0.59 |
1 |
1 |
1 |
0.6 |
0.69 |
0.29 |
0.91 |
0.47 |
sipu/jain |
0.89 |
0.72 |
0.89 |
0.16 |
0.25 |
1 |
1 |
0.57 |
0.57 |
0.62 |
0.72 |
0.72 |
sipu/pathbased |
0.66 |
0.64 |
0.52 |
0.66 |
0.98 |
0.98 |
0.74 |
0.68 |
0.62 |
0.09 |
0.68 |
0.64 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.99 |
0.99 |
0.98 |
0.99 |
0.99 |
0.99 |
0.99 |
0.8 |
0.99 |
0.44 |
0.99 |
0.99 |
sipu/s2 |
0.95 |
0.95 |
0.84 |
0.97 |
0.96 |
0.96 |
0.84 |
0.86 |
0.97 |
0.01 |
0.97 |
0.95 |
sipu/s3 |
0.67 |
0.8 |
0.57 |
0.85 |
0.82 |
0.77 |
0.63 |
0.75 |
0.84 |
0.01 |
0.84 |
0.81 |
sipu/s4 |
0.59 |
0.68 |
0.53 |
0.79 |
0.77 |
0.73 |
0.54 |
0.66 |
0.78 |
0.01 |
0.7 |
0.68 |
sipu/spiral |
0.04 |
0.07 |
0.06 |
0.03 |
1 |
1 |
1 |
0.83 |
0.01 |
1 |
0.02 |
0.07 |
sipu/unbalance |
1 |
1 |
0.64 |
1 |
0.47 |
0.59 |
0.75 |
0.41 |
1 |
0.98 |
1 |
1 |
uci/ecoli |
0.73 |
0.59 |
0.68 |
0.66 |
0.46 |
0.51 |
0.65 |
0.46 |
0.57 |
0.37 |
0.58 |
0.59 |
uci/glass |
0.25 |
0.38 |
0.38 |
0.41 |
0.27 |
0.38 |
0.38 |
0.39 |
0.45 |
0.24 |
0.39 |
0.4 |
uci/ionosphere |
0.29 |
0.44 |
0.29 |
0.64 |
0.46 |
0.46 |
0.08 |
0.3 |
0.42 |
0.29 |
0.29 |
0.44 |
uci/sonar |
0.11 |
0.02 |
0.02 |
0.08 |
0.02 |
0.02 |
0.11 |
0.08 |
0.11 |
0.08 |
0.08 |
0.02 |
uci/statlog |
0 |
0.45 |
0.17 |
0.53 |
0.74 |
0.59 |
0.51 |
0.6 |
0.43 |
0 |
nan |
0.41 |
uci/wdbc |
0.33 |
0.56 |
0.33 |
0.84 |
0.31 |
0.55 |
0.55 |
0.79 |
0.71 |
0.26 |
0.26 |
0.56 |
uci/wine |
0.42 |
0.54 |
0.51 |
0.91 |
0.57 |
0.57 |
0.3 |
0.58 |
0.55 |
0.14 |
0.44 |
0.54 |
uci/yeast |
0.25 |
0.32 |
0.28 |
0.29 |
0.24 |
0.36 |
0.3 |
0.21 |
0.32 |
0.24 |
0.25 |
0.29 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.03 |
0.17 |
0.32 |
0.48 |
0.87 |
0.68 |
0.38 |
0.44 |
0.39 |
0 |
0.01 |
0.31 |
wut/graph |
0.56 |
0.58 |
0.56 |
0.93 |
0.63 |
0.59 |
0.45 |
0.63 |
0.59 |
0.07 |
0.58 |
0.62 |
wut/isolation |
0.03 |
0.04 |
0.02 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.05 |
wut/labirynth |
0.5 |
0.59 |
0.46 |
0.65 |
0.55 |
0.61 |
0.72 |
0.72 |
0.44 |
0.72 |
0.51 |
0.46 |
wut/mk1 |
1 |
1 |
0.98 |
1 |
1 |
1 |
1 |
0.68 |
1 |
0.5 |
1 |
1 |
wut/mk2 |
0.07 |
0.11 |
0.09 |
0.09 |
1 |
1 |
1 |
1 |
0.09 |
1 |
0.08 |
0.07 |
wut/mk3 |
0.5 |
0.92 |
0.92 |
0.94 |
0.88 |
0.88 |
0.58 |
0.68 |
0.94 |
0 |
0.94 |
0.93 |
wut/mk4 |
0.18 |
0.31 |
0.42 |
0.55 |
1 |
1 |
1 |
0.74 |
0.37 |
1 |
0.39 |
0.36 |
wut/olympic |
0.27 |
0.26 |
0.21 |
0.2 |
0.32 |
0.29 |
0.25 |
0.36 |
0.22 |
0 |
0.26 |
0.21 |
wut/smile |
0.99 |
0.71 |
0.83 |
0.54 |
0.63 |
1 |
1 |
0.58 |
0.72 |
1 |
1 |
0.77 |
wut/stripes |
0.05 |
0.04 |
0.11 |
0.11 |
1 |
1 |
1 |
1 |
0.11 |
1 |
0.11 |
0.12 |
wut/trajectories |
1 |
1 |
0.74 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.25 |
0.29 |
0.5 |
0.3 |
0.57 |
1 |
1 |
0.56 |
0.36 |
1 |
0.88 |
0.37 |
wut/twosplashes |
0.05 |
0.53 |
0.47 |
0.82 |
0.71 |
0.71 |
0.71 |
0.86 |
0.53 |
0.01 |
0.53 |
0.44 |
wut/windows |
0.35 |
0.34 |
0.19 |
0.42 |
0.31 |
0.35 |
1 |
0.33 |
0.3 |
1 |
0.18 |
0.31 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.61 |
0.99 |
1 |
0.84 |
0.84 |
0.84 |
0.56 |
0.84 |
0.99 |
0.22 |
0.42 |
0.99 |
wut/x3 |
0.97 |
0.99 |
0.7 |
0.97 |
0.94 |
0.97 |
0.97 |
0.69 |
1 |
0.39 |
0.58 |
0.99 |
wut/z1 |
0.32 |
0.27 |
0.36 |
0.11 |
0.5 |
0.5 |
0.3 |
0.5 |
0.31 |
0.06 |
0.33 |
0.27 |
wut/z2 |
0.59 |
0.67 |
0.59 |
1 |
0.66 |
0.7 |
0.81 |
0.65 |
0.64 |
0.86 |
0.99 |
0.57 |
wut/z3 |
1 |
1 |
0.96 |
1 |
0.67 |
0.68 |
0.95 |
0.74 |
1 |
0.73 |
0.99 |
1 |
psi¶
Summary statistics for psi (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
73 |
0.51 |
0.37 |
0 |
0.19 |
0.45 |
0.94 |
1 |
Birch_0.01 |
73 |
0.56 |
0.34 |
0 |
0.24 |
0.53 |
0.94 |
1 |
Complete linkage |
73 |
0.49 |
0.33 |
0 |
0.26 |
0.4 |
0.75 |
1 |
Gaussian mixtures |
73 |
0.64 |
0.37 |
0 |
0.3 |
0.82 |
0.98 |
1 |
Genie_0.1 |
73 |
0.73 |
0.32 |
0 |
0.47 |
0.89 |
1 |
1 |
Genie_0.3 |
73 |
0.77 |
0.28 |
0 |
0.52 |
0.94 |
1 |
1 |
Genie_0.5 |
73 |
0.75 |
0.31 |
0 |
0.51 |
0.9 |
1 |
1 |
ITM |
73 |
0.67 |
0.28 |
0.01 |
0.49 |
0.72 |
0.99 |
1 |
K-means |
73 |
0.57 |
0.34 |
0 |
0.28 |
0.53 |
0.96 |
1 |
Single linkage |
73 |
0.41 |
0.44 |
0 |
0 |
0.2 |
1 |
1 |
Spectral_RBF_5 |
72 |
0.64 |
0.36 |
0 |
0.3 |
0.76 |
0.99 |
1 |
Ward linkage |
73 |
0.56 |
0.33 |
0 |
0.25 |
0.51 |
0.93 |
1 |
Ranks for psi (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Average linkage |
72 |
6.7 |
3.6 |
1 |
4 |
7.5 |
10 |
12 |
Birch_0.01 |
72 |
5.6 |
3.2 |
1 |
3 |
6 |
8 |
12 |
Complete linkage |
72 |
7.9 |
2.9 |
1 |
7 |
8 |
10 |
12 |
Gaussian mixtures |
72 |
4.6 |
4 |
1 |
1 |
3.5 |
9 |
12 |
Genie_0.1 |
72 |
4 |
3.5 |
1 |
1 |
3 |
7 |
12 |
Genie_0.3 |
72 |
3.6 |
3.3 |
1 |
1 |
2 |
5.2 |
12 |
Genie_0.5 |
72 |
4.3 |
4.1 |
1 |
1 |
1 |
9.2 |
12 |
ITM |
72 |
5.3 |
3.9 |
1 |
1 |
5 |
9 |
12 |
K-means |
72 |
5 |
3.5 |
1 |
1 |
5 |
8 |
12 |
Single linkage |
72 |
7.8 |
5 |
1 |
1 |
11 |
12 |
12 |
Spectral_RBF_5 |
72 |
5.4 |
3.6 |
1 |
1 |
6 |
8.2 |
11 |
Ward linkage |
72 |
5.9 |
3.3 |
1 |
3 |
6 |
8 |
12 |
Raw results for psi (best=1.0):
Average linkage |
Birch_0.01 |
Complete linkage |
Gaussian mixtures |
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Spectral_RBF_5 |
Ward linkage |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|
fcps/atom |
0.19 |
0.19 |
0.17 |
0.09 |
1 |
1 |
1 |
1 |
0.27 |
1 |
1 |
0.19 |
fcps/chainlink |
0.35 |
0.36 |
0.39 |
0.95 |
1 |
1 |
1 |
1 |
0.31 |
1 |
1 |
0.36 |
fcps/engytime |
0.13 |
0.88 |
0.11 |
0.98 |
0.85 |
0.85 |
0.85 |
0.91 |
0.88 |
0 |
0.92 |
0.77 |
fcps/hepta |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
0.9 |
1 |
1 |
1 |
1 |
fcps/lsun |
0.45 |
0.51 |
0.51 |
1 |
1 |
1 |
1 |
1 |
0.54 |
1 |
0.81 |
0.46 |
fcps/target |
0.79 |
0.25 |
0.82 |
0.17 |
1 |
1 |
1 |
1 |
0.28 |
1 |
1 |
0.33 |
fcps/tetra |
0.99 |
0.97 |
0.99 |
1 |
1 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.97 |
fcps/twodiamonds |
1 |
1 |
0.99 |
1 |
0.98 |
0.98 |
0.98 |
0.99 |
1 |
0 |
1 |
1 |
fcps/wingnut |
1 |
1 |
1 |
0.93 |
1 |
1 |
1 |
1 |
0.93 |
1 |
0.88 |
0.78 |
graves/dense |
0.92 |
0.92 |
0.34 |
1 |
0.96 |
0.96 |
0.96 |
1 |
0.82 |
0.02 |
0.92 |
0.92 |
graves/fuzzyx |
0.76 |
0.9 |
0.94 |
1 |
0.74 |
0.59 |
0.5 |
0.95 |
1 |
0 |
0.67 |
0.79 |
graves/line |
0 |
0 |
0 |
1 |
0.02 |
1 |
1 |
0.2 |
0 |
1 |
1 |
0 |
graves/parabolic |
0.75 |
0.6 |
0.67 |
0.7 |
0.84 |
0.84 |
0.84 |
0.78 |
0.76 |
0 |
0.79 |
0.72 |
graves/ring |
0.2 |
0.21 |
0.3 |
0.09 |
1 |
1 |
1 |
1 |
0.01 |
1 |
1 |
0.21 |
graves/ring_noisy |
0 |
0.2 |
0.35 |
0.08 |
1 |
1 |
1 |
1 |
0.25 |
0 |
1 |
0.24 |
graves/ring_outliers |
0.19 |
0.17 |
0.15 |
0.14 |
1 |
1 |
1 |
1 |
0.24 |
1 |
1 |
0.17 |
graves/zigzag |
0.54 |
0.64 |
0.38 |
0.96 |
1 |
1 |
1 |
0.79 |
0.6 |
1 |
0.71 |
0.53 |
graves/zigzag_noisy |
0.51 |
0.68 |
0.42 |
0.93 |
0.8 |
1 |
1 |
0.62 |
0.52 |
0.4 |
0.68 |
0.71 |
graves/zigzag_outliers |
0.17 |
0.4 |
0.29 |
0.98 |
1 |
1 |
1 |
0.83 |
0.33 |
0.4 |
0.22 |
0.25 |
other/chameleon_t4_8k |
0.54 |
0.51 |
0.48 |
0.47 |
0.64 |
1 |
0.72 |
0.69 |
0.52 |
0 |
0.52 |
0.48 |
other/chameleon_t5_8k |
1 |
1 |
0.73 |
1 |
1 |
1 |
0.71 |
0.57 |
1 |
0 |
1 |
1 |
other/chameleon_t7_10k |
0.33 |
0.28 |
0.28 |
0.27 |
0.42 |
0.71 |
1 |
0.46 |
0.31 |
0 |
0.31 |
0.37 |
other/chameleon_t8_8k |
0.28 |
0.34 |
0.26 |
0.34 |
0.41 |
0.34 |
0.68 |
0.4 |
0.28 |
0 |
0.27 |
0.28 |
other/hdbscan |
0.19 |
0.56 |
0.39 |
0.74 |
0.97 |
0.69 |
0.69 |
0.78 |
0.72 |
0 |
0.34 |
0.86 |
other/iris |
0.76 |
0.74 |
0.64 |
0.91 |
0.9 |
0.9 |
0.4 |
0.9 |
0.76 |
0.38 |
0.75 |
0.74 |
other/iris5 |
0.77 |
0.75 |
0.65 |
0.91 |
0.34 |
0.34 |
0.91 |
0.28 |
0.76 |
0.41 |
0.75 |
0.75 |
other/square |
0.24 |
0.01 |
0.26 |
0.17 |
1 |
1 |
1 |
1 |
0.15 |
1 |
0.26 |
0.34 |
sipu/a1 |
0.94 |
0.94 |
0.93 |
0.97 |
0.95 |
0.88 |
0.76 |
0.74 |
0.98 |
0.2 |
0.95 |
0.93 |
sipu/a2 |
0.95 |
0.95 |
0.92 |
0.97 |
0.96 |
0.92 |
0.75 |
0.77 |
0.98 |
0.14 |
0.9 |
0.93 |
sipu/a3 |
0.95 |
0.95 |
0.93 |
0.94 |
0.97 |
0.94 |
0.77 |
0.76 |
0.94 |
0.1 |
0.92 |
0.95 |
sipu/aggregation |
1 |
0.7 |
0.67 |
1 |
0.31 |
0.45 |
0.8 |
0.47 |
0.65 |
0.58 |
0.99 |
0.7 |
sipu/compound |
0.64 |
0.67 |
0.64 |
0.67 |
0.59 |
0.67 |
0.7 |
0.54 |
0.68 |
0.64 |
0.65 |
0.67 |
sipu/d31 |
0.91 |
0.95 |
0.95 |
0.96 |
0.95 |
0.9 |
0.69 |
0.81 |
0.97 |
0.15 |
0.95 |
0.94 |
sipu/flame |
0.48 |
0.17 |
0 |
0.36 |
1 |
1 |
1 |
0.37 |
0.56 |
0.01 |
0.91 |
0.17 |
sipu/jain |
0.74 |
0.53 |
0.74 |
0 |
0.01 |
1 |
1 |
0.39 |
0.39 |
0.21 |
0.53 |
0.53 |
sipu/pathbased |
0.4 |
0.49 |
0.35 |
0.4 |
0.97 |
0.97 |
0.6 |
0.57 |
0.42 |
0 |
0.43 |
0.49 |
sipu/r15 |
1 |
1 |
1 |
1 |
0.99 |
0.99 |
1 |
0.99 |
1 |
1 |
1 |
1 |
sipu/s1 |
0.99 |
0.99 |
0.97 |
0.99 |
0.99 |
0.99 |
0.99 |
0.73 |
0.99 |
0.28 |
0.99 |
0.99 |
sipu/s2 |
0.94 |
0.92 |
0.75 |
0.96 |
0.94 |
0.94 |
0.78 |
0.8 |
0.96 |
0 |
0.96 |
0.93 |
sipu/s3 |
0.55 |
0.77 |
0.46 |
0.82 |
0.76 |
0.7 |
0.51 |
0.69 |
0.82 |
0 |
0.8 |
0.76 |
sipu/s4 |
0.44 |
0.62 |
0.4 |
0.75 |
0.72 |
0.67 |
0.41 |
0.59 |
0.75 |
0 |
0.59 |
0.63 |
sipu/spiral |
0.03 |
0.06 |
0.05 |
0.03 |
1 |
1 |
1 |
0.72 |
0.01 |
1 |
0.02 |
0.06 |
sipu/unbalance |
1 |
1 |
0.74 |
1 |
0.17 |
0.21 |
0.26 |
0.15 |
1 |
0.78 |
1 |
1 |
uci/ecoli |
0.39 |
0.41 |
0.34 |
0.39 |
0.29 |
0.3 |
0.27 |
0.33 |
0.4 |
0.18 |
0.34 |
0.41 |
uci/glass |
0.06 |
0.21 |
0.21 |
0.22 |
0.19 |
0.27 |
0.21 |
0.22 |
0.27 |
0.05 |
0.16 |
0.22 |
uci/ionosphere |
0.01 |
0.35 |
0.01 |
0.53 |
0.4 |
0.4 |
0 |
0.17 |
0.34 |
0.01 |
0.01 |
0.35 |
uci/sonar |
0.03 |
0 |
0 |
0.01 |
0 |
0 |
0.04 |
0.01 |
0.05 |
0.01 |
0.01 |
0 |
uci/statlog |
0 |
0.35 |
0.13 |
0.42 |
0.71 |
0.52 |
0.42 |
0.52 |
0.32 |
0 |
nan |
0.32 |
uci/wdbc |
0.06 |
0.3 |
0.06 |
0.73 |
0.06 |
0.3 |
0.3 |
0.73 |
0.5 |
0 |
0 |
0.3 |
uci/wine |
0.23 |
0.48 |
0.42 |
0.83 |
0.46 |
0.46 |
0.13 |
0.46 |
0.48 |
0.03 |
0.25 |
0.48 |
uci/yeast |
0.17 |
0.23 |
0.17 |
0.18 |
0.16 |
0.17 |
0.13 |
0.13 |
0.24 |
0.13 |
0.14 |
0.21 |
wut/circles |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/cross |
0.02 |
0.11 |
0.17 |
0.3 |
0.78 |
0.55 |
0.27 |
0.33 |
0.28 |
0 |
0.01 |
0.22 |
wut/graph |
0.38 |
0.42 |
0.38 |
0.88 |
0.53 |
0.46 |
0.26 |
0.55 |
0.43 |
0.05 |
0.4 |
0.46 |
wut/isolation |
0.03 |
0.03 |
0.02 |
0.01 |
1 |
1 |
1 |
1 |
0.01 |
1 |
0.01 |
0.04 |
wut/labirynth |
0.31 |
0.42 |
0.31 |
0.57 |
0.35 |
0.46 |
0.69 |
0.69 |
0.31 |
0.52 |
0.34 |
0.29 |
wut/mk1 |
0.99 |
0.99 |
0.97 |
0.99 |
0.99 |
0.99 |
0.99 |
0.55 |
0.99 |
0.35 |
0.99 |
0.99 |
wut/mk2 |
0.07 |
0.09 |
0.09 |
0.08 |
1 |
1 |
1 |
1 |
0.09 |
1 |
0.07 |
0.06 |
wut/mk3 |
0.35 |
0.89 |
0.87 |
0.92 |
0.82 |
0.82 |
0.43 |
0.56 |
0.93 |
0 |
0.92 |
0.91 |
wut/mk4 |
0.1 |
0.2 |
0.3 |
0.55 |
1 |
1 |
1 |
0.63 |
0.25 |
1 |
0.27 |
0.24 |
wut/olympic |
0.22 |
0.24 |
0.2 |
0.18 |
0.25 |
0.26 |
0.21 |
0.31 |
0.21 |
0 |
0.24 |
0.19 |
wut/smile |
0.98 |
0.65 |
0.58 |
0.43 |
0.47 |
1 |
1 |
0.62 |
0.66 |
1 |
1 |
0.69 |
wut/stripes |
0.04 |
0.03 |
0.09 |
0.11 |
1 |
1 |
1 |
1 |
0.1 |
1 |
0.11 |
0.08 |
wut/trajectories |
1 |
1 |
0.62 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/trapped_lovers |
0.13 |
0.15 |
0.34 |
0.17 |
0.5 |
1 |
1 |
0.49 |
0.21 |
1 |
0.77 |
0.21 |
wut/twosplashes |
0.03 |
0.53 |
0.44 |
0.82 |
0.71 |
0.71 |
0.71 |
0.75 |
0.53 |
0.01 |
0.53 |
0.37 |
wut/windows |
0.28 |
0.28 |
0.06 |
0.34 |
0.29 |
0.39 |
1 |
0.2 |
0.24 |
1 |
0.08 |
0.26 |
wut/x1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
wut/x2 |
0.56 |
0.98 |
1 |
0.74 |
0.74 |
0.74 |
0.5 |
0.75 |
0.98 |
0 |
0.19 |
0.98 |
wut/x3 |
0.92 |
0.98 |
0.59 |
0.94 |
0.89 |
0.92 |
0.92 |
0.49 |
1 |
0.02 |
0.45 |
0.98 |
wut/z1 |
0.25 |
0.2 |
0.33 |
0.09 |
0.38 |
0.38 |
0.19 |
0.38 |
0.31 |
0.04 |
0.31 |
0.2 |
wut/z2 |
0.45 |
0.56 |
0.39 |
1 |
0.56 |
0.47 |
0.63 |
0.56 |
0.51 |
0.47 |
0.99 |
0.51 |
wut/z3 |
1 |
0.99 |
0.92 |
0.99 |
0.49 |
0.44 |
0.9 |
0.6 |
1 |
0.55 |
0.97 |
1 |
Summary¶
Medians and means of the partition similarity scores (read row-wise, in groups of 2 columns):
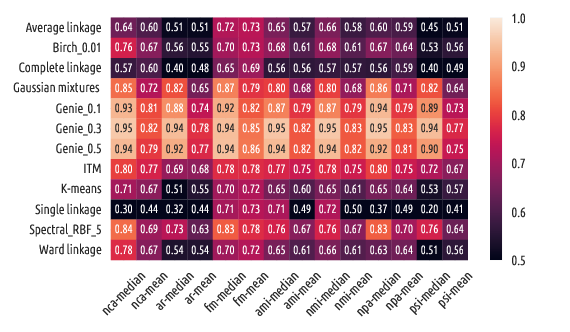
Figure 22 Heat map of median and mean similarity scores¶
Large Datase¶
Results¶
nca¶
Summary statistics for nca (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.75 |
0.22 |
0.43 |
0.6 |
0.76 |
0.92 |
1 |
Genie_0.3 |
6 |
0.61 |
0.33 |
0.16 |
0.38 |
0.6 |
0.88 |
1 |
Genie_0.5 |
6 |
0.39 |
0.43 |
0 |
0.06 |
0.25 |
0.71 |
1 |
ITM |
6 |
0.71 |
0.15 |
0.52 |
0.6 |
0.75 |
0.79 |
0.9 |
K-means |
6 |
0.68 |
0.26 |
0.42 |
0.47 |
0.62 |
0.91 |
0.99 |
Single linkage |
6 |
0.1 |
0.24 |
0 |
0 |
0 |
0 |
0.59 |
Ward linkage |
6 |
0.69 |
0.23 |
0.44 |
0.5 |
0.65 |
0.86 |
1 |
Ranks for nca (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.7 |
0.8 |
1 |
1 |
1.5 |
2 |
3 |
Genie_0.3 |
6 |
3.5 |
1.8 |
1 |
2.2 |
4 |
5 |
5 |
Genie_0.5 |
6 |
5 |
2 |
1 |
5.2 |
6 |
6 |
6 |
ITM |
6 |
3 |
2.1 |
1 |
1.2 |
2.5 |
4.5 |
6 |
K-means |
6 |
3.5 |
1.6 |
1 |
2.5 |
4 |
4.8 |
5 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
3 |
1.3 |
1 |
2.2 |
3.5 |
4 |
4 |
Raw results for nca (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.69 |
0.34 |
0.06 |
0.9 |
0.48 |
0 |
0.58 |
mnist/fashion |
0.43 |
0.16 |
0.06 |
0.56 |
0.42 |
0 |
0.44 |
sipu/birch1 |
0.94 |
0.94 |
0.8 |
0.8 |
0.96 |
0 |
0.91 |
sipu/birch2 |
1 |
1 |
1 |
0.77 |
0.99 |
0.59 |
1 |
sipu/worms_2 |
0.57 |
0.49 |
0.44 |
0.52 |
0.46 |
0 |
0.47 |
sipu/worms_64 |
0.84 |
0.7 |
0 |
0.74 |
0.75 |
0 |
0.73 |
ar¶
Summary statistics for ar (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.66 |
0.26 |
0.31 |
0.48 |
0.65 |
0.85 |
1 |
Genie_0.3 |
6 |
0.52 |
0.37 |
0.07 |
0.25 |
0.46 |
0.81 |
1 |
Genie_0.5 |
6 |
0.35 |
0.43 |
0 |
0.02 |
0.17 |
0.63 |
1 |
ITM |
6 |
0.6 |
0.18 |
0.37 |
0.46 |
0.65 |
0.71 |
0.8 |
K-means |
6 |
0.61 |
0.31 |
0.32 |
0.35 |
0.52 |
0.88 |
0.99 |
Single linkage |
6 |
0.07 |
0.18 |
0 |
0 |
0 |
0 |
0.44 |
Ward linkage |
6 |
0.58 |
0.27 |
0.33 |
0.38 |
0.47 |
0.75 |
1 |
Ranks for ar (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.8 |
1.2 |
1 |
1 |
1.5 |
2 |
4 |
Genie_0.3 |
6 |
3.2 |
1.7 |
1 |
2 |
3 |
4.8 |
5 |
Genie_0.5 |
6 |
4.8 |
1.9 |
1 |
5 |
5.5 |
6 |
6 |
ITM |
6 |
3.3 |
2.3 |
1 |
1.5 |
3 |
5.2 |
6 |
K-means |
6 |
3.3 |
1.6 |
1 |
2.2 |
3.5 |
4.8 |
5 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
3.2 |
1.5 |
1 |
2.2 |
3.5 |
4 |
5 |
Raw results for ar (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.6 |
0.21 |
0.02 |
0.8 |
0.37 |
0 |
0.53 |
mnist/fashion |
0.31 |
0.07 |
0.02 |
0.41 |
0.35 |
0 |
0.37 |
sipu/birch1 |
0.89 |
0.89 |
0.73 |
0.72 |
0.94 |
0 |
0.83 |
sipu/birch2 |
1 |
1 |
1 |
0.71 |
0.99 |
0.44 |
1 |
sipu/worms_2 |
0.45 |
0.38 |
0.32 |
0.37 |
0.32 |
0 |
0.33 |
sipu/worms_64 |
0.7 |
0.54 |
0 |
0.59 |
0.67 |
0 |
0.42 |
fm¶
Summary statistics for fm (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.69 |
0.23 |
0.42 |
0.51 |
0.68 |
0.85 |
1 |
Genie_0.3 |
6 |
0.6 |
0.29 |
0.31 |
0.4 |
0.49 |
0.82 |
1 |
Genie_0.5 |
6 |
0.49 |
0.31 |
0.2 |
0.31 |
0.34 |
0.65 |
1 |
ITM |
6 |
0.62 |
0.16 |
0.39 |
0.51 |
0.66 |
0.72 |
0.82 |
K-means |
6 |
0.64 |
0.28 |
0.35 |
0.42 |
0.56 |
0.88 |
0.99 |
Single linkage |
6 |
0.28 |
0.15 |
0.1 |
0.19 |
0.26 |
0.32 |
0.53 |
Ward linkage |
6 |
0.61 |
0.25 |
0.36 |
0.45 |
0.52 |
0.77 |
1 |
Ranks for fm (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.7 |
0.8 |
1 |
1 |
1.5 |
2 |
3 |
Genie_0.3 |
6 |
3.3 |
2 |
1 |
2 |
3 |
4.8 |
6 |
Genie_0.5 |
6 |
4.8 |
2.1 |
1 |
4.2 |
5.5 |
6 |
7 |
ITM |
6 |
3.3 |
2.3 |
1 |
1.5 |
3 |
5.2 |
6 |
K-means |
6 |
3.5 |
1.9 |
1 |
2.2 |
3.5 |
4.8 |
6 |
Single linkage |
6 |
6.3 |
0.8 |
5 |
6 |
6.5 |
7 |
7 |
Ward linkage |
6 |
3.2 |
1.5 |
1 |
2.2 |
3.5 |
4 |
5 |
Raw results for fm (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.65 |
0.41 |
0.31 |
0.82 |
0.43 |
0.32 |
0.58 |
mnist/fashion |
0.42 |
0.31 |
0.31 |
0.47 |
0.42 |
0.32 |
0.44 |
sipu/birch1 |
0.89 |
0.89 |
0.75 |
0.72 |
0.94 |
0.1 |
0.83 |
sipu/birch2 |
1 |
1 |
1 |
0.72 |
0.99 |
0.53 |
1 |
sipu/worms_2 |
0.47 |
0.4 |
0.36 |
0.39 |
0.35 |
0.19 |
0.36 |
sipu/worms_64 |
0.72 |
0.58 |
0.2 |
0.61 |
0.69 |
0.2 |
0.47 |
ami¶
Summary statistics for ami (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.77 |
0.17 |
0.57 |
0.67 |
0.74 |
0.9 |
1 |
Genie_0.3 |
6 |
0.7 |
0.24 |
0.37 |
0.59 |
0.66 |
0.88 |
1 |
Genie_0.5 |
6 |
0.49 |
0.42 |
0 |
0.19 |
0.41 |
0.84 |
1 |
ITM |
6 |
0.75 |
0.15 |
0.56 |
0.63 |
0.74 |
0.87 |
0.91 |
K-means |
6 |
0.72 |
0.22 |
0.5 |
0.53 |
0.66 |
0.91 |
1 |
Single linkage |
6 |
0.15 |
0.36 |
0 |
0 |
0 |
0 |
0.89 |
Ward linkage |
6 |
0.75 |
0.17 |
0.56 |
0.62 |
0.7 |
0.87 |
1 |
Ranks for ami (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.3 |
0.5 |
1 |
1 |
1 |
1.8 |
2 |
Genie_0.3 |
6 |
3 |
1.5 |
1 |
2 |
3 |
4 |
5 |
Genie_0.5 |
6 |
4.5 |
2 |
1 |
4 |
5 |
6 |
6 |
ITM |
6 |
3.8 |
2.1 |
1 |
2.2 |
4 |
5.8 |
6 |
K-means |
6 |
2.8 |
2 |
1 |
1 |
2.5 |
4.8 |
5 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
2.7 |
1.6 |
1 |
1.2 |
2.5 |
3.8 |
5 |
Raw results for ami (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.75 |
0.57 |
0.18 |
0.83 |
0.5 |
0 |
0.68 |
mnist/fashion |
0.57 |
0.37 |
0.21 |
0.56 |
0.51 |
0 |
0.56 |
sipu/birch1 |
0.94 |
0.94 |
0.92 |
0.89 |
0.98 |
0 |
0.92 |
sipu/birch2 |
1 |
1 |
1 |
0.91 |
1 |
0.89 |
1 |
sipu/worms_2 |
0.65 |
0.63 |
0.61 |
0.62 |
0.6 |
0 |
0.6 |
sipu/worms_64 |
0.72 |
0.68 |
0 |
0.66 |
0.72 |
0 |
0.72 |
nmi¶
Summary statistics for nmi (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.77 |
0.17 |
0.57 |
0.67 |
0.74 |
0.9 |
1 |
Genie_0.3 |
6 |
0.7 |
0.24 |
0.37 |
0.59 |
0.66 |
0.88 |
1 |
Genie_0.5 |
6 |
0.49 |
0.42 |
0 |
0.19 |
0.41 |
0.84 |
1 |
ITM |
6 |
0.75 |
0.15 |
0.56 |
0.63 |
0.75 |
0.87 |
0.91 |
K-means |
6 |
0.72 |
0.22 |
0.5 |
0.53 |
0.66 |
0.91 |
1 |
Single linkage |
6 |
0.15 |
0.36 |
0 |
0 |
0 |
0 |
0.89 |
Ward linkage |
6 |
0.75 |
0.17 |
0.56 |
0.62 |
0.7 |
0.87 |
1 |
Ranks for nmi (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.5 |
0.5 |
1 |
1 |
1.5 |
2 |
2 |
Genie_0.3 |
6 |
3 |
1.5 |
1 |
2 |
3 |
4 |
5 |
Genie_0.5 |
6 |
4.5 |
2 |
1 |
4 |
5 |
6 |
6 |
ITM |
6 |
3.8 |
2.1 |
1 |
2.2 |
4 |
5.8 |
6 |
K-means |
6 |
3 |
1.9 |
1 |
1.2 |
3 |
4.8 |
5 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
2.7 |
1.6 |
1 |
1.2 |
2.5 |
3.8 |
5 |
Raw results for nmi (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.75 |
0.57 |
0.18 |
0.83 |
0.5 |
0 |
0.68 |
mnist/fashion |
0.57 |
0.37 |
0.21 |
0.56 |
0.51 |
0 |
0.56 |
sipu/birch1 |
0.94 |
0.94 |
0.92 |
0.89 |
0.98 |
0 |
0.92 |
sipu/birch2 |
1 |
1 |
1 |
0.91 |
1 |
0.89 |
1 |
sipu/worms_2 |
0.65 |
0.63 |
0.61 |
0.62 |
0.6 |
0 |
0.6 |
sipu/worms_64 |
0.72 |
0.68 |
0 |
0.66 |
0.72 |
0 |
0.73 |
npa¶
Summary statistics for npa (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.75 |
0.22 |
0.43 |
0.61 |
0.77 |
0.92 |
1 |
Genie_0.3 |
6 |
0.61 |
0.33 |
0.16 |
0.39 |
0.61 |
0.88 |
1 |
Genie_0.5 |
6 |
0.4 |
0.43 |
0 |
0.06 |
0.26 |
0.71 |
1 |
ITM |
6 |
0.71 |
0.15 |
0.49 |
0.6 |
0.75 |
0.79 |
0.89 |
K-means |
6 |
0.67 |
0.26 |
0.42 |
0.44 |
0.62 |
0.91 |
0.99 |
Single linkage |
6 |
0.1 |
0.24 |
0 |
0 |
0.01 |
0.02 |
0.59 |
Ward linkage |
6 |
0.68 |
0.23 |
0.44 |
0.48 |
0.65 |
0.86 |
1 |
Ranks for npa (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.7 |
0.8 |
1 |
1 |
1.5 |
2 |
3 |
Genie_0.3 |
6 |
3.3 |
1.9 |
1 |
2 |
3.5 |
5 |
5 |
Genie_0.5 |
6 |
4.8 |
1.9 |
1 |
5 |
5.5 |
6 |
6 |
ITM |
6 |
3.2 |
2 |
1 |
1.5 |
3 |
4.5 |
6 |
K-means |
6 |
3.7 |
1.9 |
1 |
2.5 |
4 |
4.8 |
6 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
3 |
1.3 |
1 |
2.2 |
3.5 |
4 |
4 |
Raw results for npa (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.7 |
0.35 |
0.07 |
0.89 |
0.48 |
0.01 |
0.58 |
mnist/fashion |
0.43 |
0.16 |
0.06 |
0.56 |
0.42 |
0 |
0.44 |
sipu/birch1 |
0.94 |
0.94 |
0.8 |
0.8 |
0.96 |
0 |
0.91 |
sipu/birch2 |
1 |
1 |
1 |
0.77 |
0.99 |
0.59 |
1 |
sipu/worms_2 |
0.58 |
0.51 |
0.44 |
0.49 |
0.43 |
0.03 |
0.45 |
sipu/worms_64 |
0.84 |
0.7 |
0 |
0.74 |
0.75 |
0 |
0.73 |
psi¶
Summary statistics for psi (best=1.0):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
0.7 |
0.25 |
0.37 |
0.51 |
0.72 |
0.9 |
1 |
Genie_0.3 |
6 |
0.57 |
0.35 |
0.13 |
0.31 |
0.53 |
0.86 |
1 |
Genie_0.5 |
6 |
0.36 |
0.42 |
0 |
0.04 |
0.18 |
0.63 |
1 |
ITM |
6 |
0.65 |
0.16 |
0.39 |
0.57 |
0.69 |
0.73 |
0.85 |
K-means |
6 |
0.62 |
0.3 |
0.31 |
0.36 |
0.57 |
0.88 |
0.98 |
Single linkage |
6 |
0.09 |
0.21 |
0 |
0 |
0 |
0 |
0.52 |
Ward linkage |
6 |
0.62 |
0.28 |
0.31 |
0.39 |
0.6 |
0.83 |
1 |
Ranks for psi (best=1):
count |
mean |
std |
min |
25% |
50% |
75% |
max |
|
|---|---|---|---|---|---|---|---|---|
Genie_0.1 |
6 |
1.5 |
0.5 |
1 |
1 |
1.5 |
2 |
2 |
Genie_0.3 |
6 |
3.3 |
1.9 |
1 |
2 |
3.5 |
5 |
5 |
Genie_0.5 |
6 |
4.8 |
2 |
1 |
4.5 |
6 |
6 |
6 |
ITM |
6 |
3.2 |
2 |
1 |
1.5 |
3 |
4.5 |
6 |
K-means |
6 |
3.7 |
1.5 |
1 |
3.2 |
4 |
4.8 |
5 |
Single linkage |
6 |
6.8 |
0.4 |
6 |
7 |
7 |
7 |
7 |
Ward linkage |
6 |
3 |
1.4 |
1 |
2.2 |
3 |
3.8 |
5 |
Raw results for psi (best=1.0):
Genie_0.1 |
Genie_0.3 |
Genie_0.5 |
ITM |
K-means |
Single linkage |
Ward linkage |
|
|---|---|---|---|---|---|---|---|
mnist/digits |
0.6 |
0.28 |
0.05 |
0.85 |
0.43 |
0 |
0.5 |
mnist/fashion |
0.37 |
0.13 |
0.04 |
0.53 |
0.33 |
0 |
0.35 |
sipu/birch1 |
0.92 |
0.92 |
0.73 |
0.75 |
0.94 |
0 |
0.87 |
sipu/birch2 |
1 |
1 |
1 |
0.69 |
0.98 |
0.52 |
1 |
sipu/worms_2 |
0.48 |
0.4 |
0.32 |
0.39 |
0.31 |
0 |
0.31 |
sipu/worms_64 |
0.84 |
0.66 |
0 |
0.7 |
0.7 |
0 |
0.71 |
Summary¶
Medians and means of the partition similarity scores:

Figure 23 Heat map of median and mean similarity scores¶