genieclust: Fast and Robust Hierarchical Clustering with Noise Point Detection¶
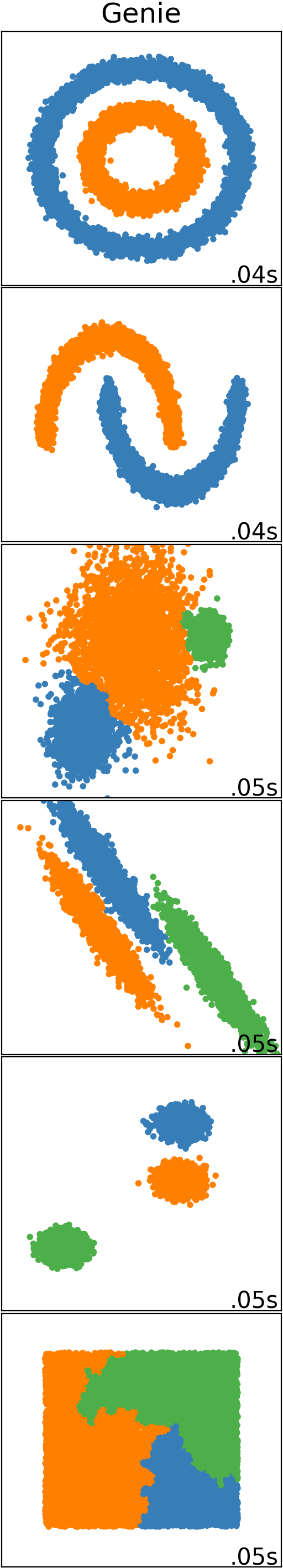
Genie finds meaningful clusters. It does so quickly, even in large datasets.
The genieclust package [4] for Python and R implements a robust and outlier-resistant hierarchical clustering algorithm called Genie [9].
The idea behind Genie is beautifully simple. First, make each individual point the only member of its own cluster. Then, keep merging pairs of the closest clusters, one after another. However, to prevent the formation of clusters of highly imbalanced sizes, a point group of the smallest size is sometimes combined with its nearest counterpart.
Genie’s appealing simplicity goes hand in hand with its usability. It often outperforms other clustering approaches such as K-means, BIRCH, or average, complete, and Ward’s linkage on benchmark datasets. Of course, there is no, nor will there ever be, a single best universal clustering approach for every kind of problem, but Genie is definitely worth a try.
Genie is based on minimal spanning trees [11] of pairwise distance graphs. Thus, it can also be pretty fast: thanks to quitefastmst, determining the entire cluster hierarchy for datasets containing millions of points can be completed in minutes. Therefore, it is well suited to solving extreme clustering tasks (involving large datasets with a high number of clusters to detect).
genieclust allows clustering with respect to mutual reachability distances,
enabling it to act as a noise point detector or a version
of HDBSCAN* [2] that can identify a predefined
number of clusters or their entire hierarchy. The good news is that it
doesn’t depend on DBSCAN’s somewhat difficult-to-set eps parameter.
Python Version¶
The Python version of genieclust is available via PyPI, e.g., via a call to:
pip3 install genieclust
from the command line or through your favourite package manager. Note the scikit-learn-like [1] API:
import genieclust
X = ... # some data
g = genieclust.Genie(n_clusters=2)
labels = g.fit_predict(X)
Note
To learn more about Python, check out Marek’s recent open-access (free!) textbook Minimalist Data Wrangling in Python [7].
R Version¶
The R version of genieclust can be downloaded from CRAN by calling:
install.packages("genieclust")
Its interface is compatible with the classic stats::hclust(),
but there is more:
X <- ... # some data
h <- gclust(X)
plot(h) # plot cluster dendrogram
cutree(h, k=2)
# or simply: genie(X, k=2)
Note
To learn more about R, check out Marek’s recent open-access (free!) textbook Deep R Programming [6].
Package Features¶
The implemented algorithms include:
Genie – a reimplementation of the original Genie algorithm from the R package genie [9]; much faster than the original one; supports arbitrary spanning forests;
Genie+HDBSCAN* – a robustified (Geniefied) retake on the HDBSCAN* [2] method that detects noise points in data and outputs clusters of predefined sizes.
Other:
inequality measures: the normalised Gini, Bonferroni, and de Vergottini indices;
external cluster validity measures [5, 8]: normalised clustering accuracy (NCA) and partition similarity scores such as normalised pivoted accuracy (NPA), adjusted/unadjusted Rand (AR), adjusted/unadjusted Fowlkes–Mallows (FM), adjusted/normalised/unadjusted mutual information (MI) indices;
internal cluster validity measures [10]: the Caliński–Harabasz, Silhouette, Ball–Hall, Davies–Bouldin, generalised Dunn indices’
(Python only) union-find (disjoint sets) data structures (with extensions);
(Python only) some R-like plotting functions.
Contributing¶
genieclust is distributed under the open source GNU AGPL v3 license and can be downloaded from GitHub. The core functionality is implemented in the form of a header-only C++ library. It can thus be easily adapted for use in other projects. New contributions are welcome, e.g., Julia, Matlab/GNU Octave wrappers.
Author and Maintainer: Marek Gagolewski
Contributors: Maciej Bartoszuk and Anna Cena (genieclust’s predecessor R package genie [9] and some internal cluster validity measures CVI [10]); Peter M. Larsen (an implementation of the shortest augmenting path algorithm for the rectangular assignment problem which we use for computing some of the external cluster validity measures [8]).