genieclust.plots¶
Various plotting functions
- genieclust.plots.plot_scatter(X, y=None, labels=None, axis=None, title=None, xlabel=None, ylabel=None, xlim=None, ylim=None, **kwargs)¶
Draws a scatter plot
- Parameters:
- Xarray_like
Either a two-column matrix that gives the x and y coordinates of the points or a vector of length n. In the latter case, y must be a vector of length n as well.
- yNone or array_like
The y coordinates of the n points in the case where X is a vector.
- labelsNone or array_like
A vector of n integer labels that correspond to each point in X, that gives its plot style.
- axis, title, xlabel, ylabel, xlim, ylimNone or object
If not None, values passed to matplotlib.pyplot.axis, matplotlib.pyplot.title, etc.
- **kwargsCollection properties
Further arguments to matplotlib.pyplot.scatter.
Notes
If X is a two-column matrix, then
plot_scatter(X)is equivalent toplot_scatter(X[:,0], X[:,1]).Unlike in matplotlib.pyplot.scatter, for any fixed
j, all pointsX[i,:]such thatlabels[i] == jare always drawn in the same way, no matter themax(labels). In particular, labels 0, 1, 2, and 3 correspond to black, red, green, and blue, respectively.This function was inspired by the
plot()function from the R packagegraphics.Examples
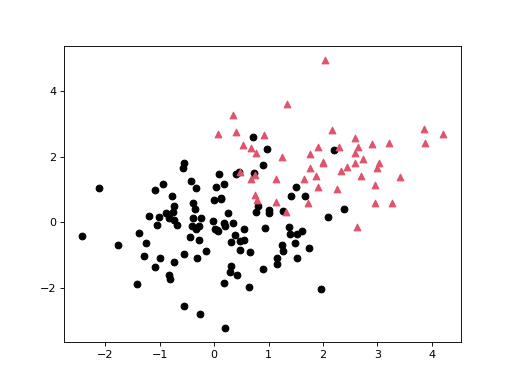
An example scatter plots where each point is assigned one of two distinct labels:
>>> n = np.r_[100, 50] >>> X = np.r_[np.random.randn(n[0], 2), np.random.randn(n[1], 2)+2.0] >>> l = np.repeat([0, 1], n) >>> genieclust.plots.plot_scatter(X, labels=l) >>> plt.show()
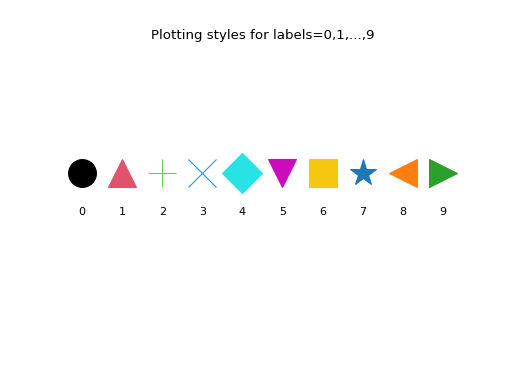
Here are the first 10 plotting styles:
>>> ncol = len(genieclust.plots.col) >>> nmrk = len(genieclust.plots.mrk) >>> mrk_recycled = np.tile( ... genieclust.plots.mrk, ... int(np.ceil(ncol/nmrk)))[:ncol] >>> for i in range(10): ... plt.text( ... i, 0, i, horizontalalignment="center") ... plt.plot( ... i, 1, marker=mrk_recycled[i], ... color=genieclust.plots.col[i], ... markersize=25) >>> plt.title("Plotting styles for labels=0,1,...,9") >>> plt.ylim(-3,4) >>> plt.axis("off") >>> plt.show()
- genieclust.plots.plot_segments(pairs, X, y=None, style='k-', **kwargs)¶
Draws a set of disjoint line segments
- Parameters:
- pairsarray_like
A two-column matrix that gives the pairs of indices defining the line segments to draw.
- Xarray_like
Either a two-column matrix that gives the x and y coordinates of the points or a vector of length n. In the latter case, y must be a vector of length n as well.
- yNone or array_like
The y coordinates of the n points in the case where X is a vector.
- style:
See matplotlib.pyplot.plot.
- **kwargsCollection properties
Further arguments to matplotlib.pyplot.plot.
Notes
The function draws a set of disjoint line segments from
(X[pairs[i,0],0], X[pairs[i,0],1])to(X[pairs[i,1],0], X[pairs[i,1],1])for allifrom0topairs.shape[0]-1.matplotlib.pyplot.plot is called only once. Therefore, you can expect it to be pretty pretty fast.
Examples
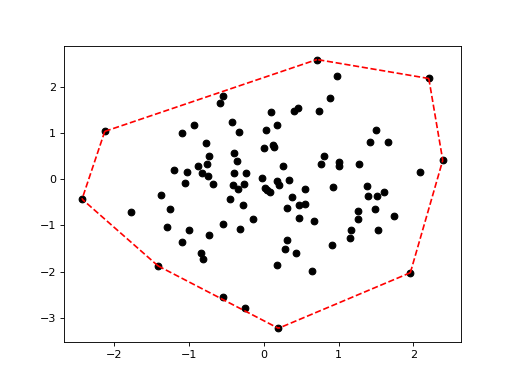
Plotting the convex hull of a point set:
>>> import scipy.spatial >>> X = np.random.randn(100, 2) >>> hull = scipy.spatial.ConvexHull(X) >>> genieclust.plots.plot_scatter(X) >>> genieclust.plots.plot_segments(hull.simplices, X, style="r--") >>> plt.show()
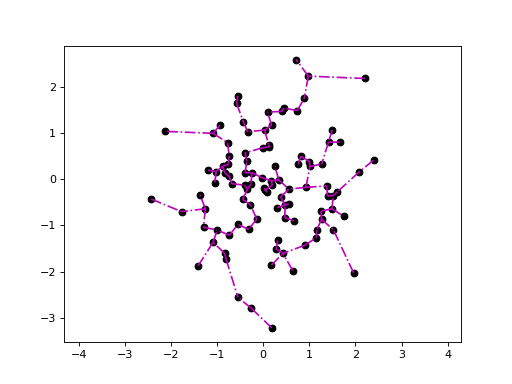
Plotting the minimum spanning tree:
>>> X = np.random.randn(100, 2) >>> mst = genieclust.internal.mst_from_distance(X, "euclidean") >>> genieclust.plots.plot_scatter(X) >>> genieclust.plots.plot_segments(mst[1], X, style="m-.") >>> plt.axis("equal") >>> plt.show()