Basic Usage (R)¶
The latest stable release of the R package genieclust is available from the CRAN repository. Install it by calling:
install.packages("genieclust")
Below are a few basic examples of interacting with the package.
library("genieclust")
Let’s take the Sustainable Society Indices dataset that measures the Human, Environmental, and Economic Wellbeing in each country. There are seven categories on the scale \([0, 10]\).
# see https://github.com/gagolews/genieclust/tree/master/devel/sphinx/weave
ssi <- read.csv("ssi_2016_categories.csv", comment.char="#")
X <- as.matrix(ssi[,-1]) # everything except the Country (1st) column
dimnames(X)[[1]] <- ssi[,1] # set row names
head(X) # preview
## BasicNeeds PersonalDevelopmentAndHealth WellBalancedSociety
## Albania 9.6058 7.9596 6.9926
## Algeria 9.0212 7.3365 4.2039
## Angola 5.9728 5.6928 2.1401
## Argentina 9.8320 8.3506 3.8952
## Armenia 9.4469 7.4205 6.2892
## Australia 10.0000 8.5909 6.1055
## NaturalResources ClimateAndEnergy Transition Economy
## Albania 6.6343 4.6217 2.1025 3.0565
## Algeria 5.2772 2.6627 3.0741 6.1543
## Angola 6.7594 6.2122 1.8988 3.7535
## Argentina 5.4535 3.3003 6.3899 5.3406
## Armenia 6.4363 2.8543 2.4342 3.8296
## Australia 4.1307 1.6278 7.5395 7.5931
The API of genieclust is compatible with R’s workhorse
for hierarchical clustering, stats::hclust(), which accepts
a complete pairwise distance matrix. Yet, for better performance,
it is better to feed genieclust::gclust() with the input data matrix
directly:
# faster than gclust(dist(X)):
h <- gclust(X) # default: gini_threshold=0.3, distance="euclidean"
print(h)
##
## Call:
## gclust.mst(d = tree, gini_threshold = gini_threshold, verbose = verbose)
##
## Cluster method : Genie(0.3)
## Distance : euclidean
## Number of objects: 154
In order to extract a desired k-partition, we can call stats::cutree():
y_pred <- cutree(h, k=3) # also: genie(X, 3)
sample(y_pred, 25) # preview
## Iran Slovenia Morocco
## 1 3 1
## Trinidad and Tobago Sierra Leone Colombia
## 1 2 1
## Turkmenistan Syria Nigeria
## 1 1 2
## Tunisia Ghana Sweden
## 1 2 1
## Netherlands Bhutan Mexico
## 3 1 1
## Jamaica Saudi Arabia Hungary
## 1 1 3
## Peru Kazakhstan Spain
## 1 1 3
## Japan Korea, North China
## 1 1 1
## Liberia
## 2
This gives the cluster IDs allocated to each country.
Note
If we are only interested in the partition of a specific cardinality,
calling genie() directly will be slightly faster than referring to
cutree(gclust(...)).
Let’s depict the obtained partition using the rworldmap package:
library("rworldmap") # see the package's manual for details
mapdata <- data.frame(Country=dimnames(X)[[1]], Cluster=y_pred)
mapdata <- joinCountryData2Map(mapdata, joinCode="NAME", nameJoinColumn="Country")
mapCountryData(mapdata, nameColumnToPlot="Cluster", catMethod="categorical",
missingCountryCol="white", colourPalette=palette.colors(3, "R4"),
mapTitle="")
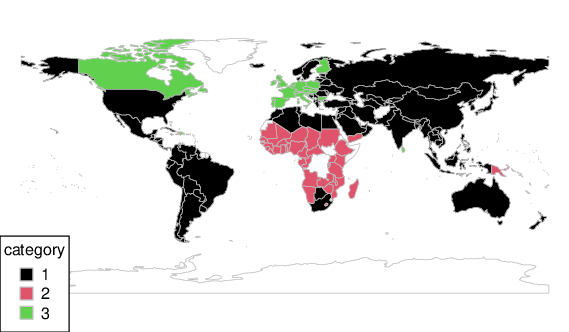
Figure 15 Countries grouped with respect to the SSI categories¶
We can compute, for instance, the average indicators in each identified group:
t(aggregate(as.data.frame(X), list(Cluster=y_pred), mean))[-1, ]
## [,1] [,2] [,3]
## BasicNeeds 9.0679 5.2689 9.8178
## PersonalDevelopmentAndHealth 7.5081 5.9312 8.2995
## WellBalancedSociety 4.8869 2.8682 6.8272
## NaturalResources 5.6633 7.0040 6.3743
## ClimateAndEnergy 3.6241 7.0818 3.5947
## Transition 4.0749 2.6300 7.3402
## Economy 5.5127 3.5411 4.2742
Dendrograms¶
Dendrogram plotting is also possible. For better readability, we will restrict ourselves to a smaller sample; namely, to the 37 members of the OECD:
oecd <- c("Australia", "Austria", "Belgium", "Canada", "Chile", "Colombia",
"Czech Republic", "Denmark", "Estonia", "Finland", "France", "Germany",
"Greece", "Hungary", "Iceland", "Ireland", "Israel", "Italy", "Japan",
"Korea, South", "Latvia", "Lithuania", "Luxembourg", "Mexico", "Netherlands",
"New Zealand", "Norway", "Poland", "Portugal", "Slovak Republic", "Slovenia",
"Spain", "Sweden", "Switzerland", "Turkey", "United Kingdom", "United States")
X_oecd <- X[dimnames(X)[[1]] %in% oecd, ]
h_oecd <- gclust(X_oecd)
plot(as.dendrogram(h_oecd), horiz=TRUE)
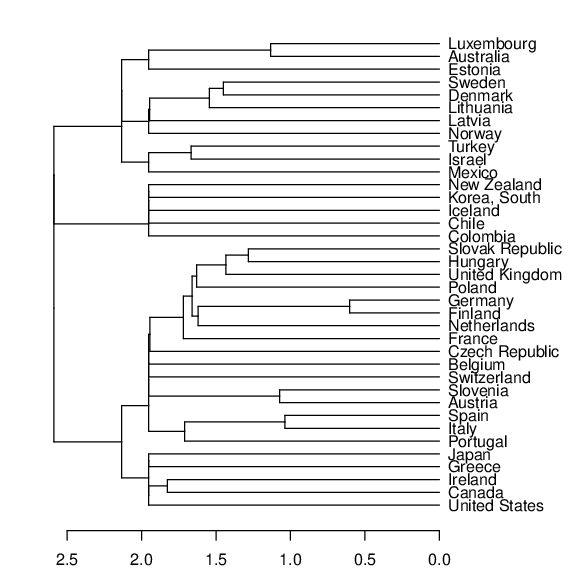
Figure 16 Cluster dendrogram for the OECD countries¶
Outlier Detection with Deadwood¶
The Deadwood outlier detection algorithm can be run on the clustered dataset to identify anomalous points in each cluster. See the deadwood package tutorials for more details.
Remarks¶
genieclust also features partition similarity scores (such as the adjusted Rand index) and internal cluster validity measures (e.g., the Caliński-Harabasz index).
For more details, refer to the package’s documentation.
To learn more about R, check out Marek’s open-access textbook Deep R Programming [6].